- Record: found
- Abstract: found
- Article: not found
Retinoblastoma and Its Binding Partner MSI1 Control Imprinting in Arabidopsis
Read this article at
Abstract
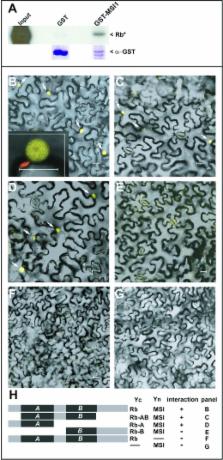
Parental genomic imprinting causes preferential expression of one of the two parental alleles. In mammals, differential sex-dependent deposition of silencing DNA methylation marks during gametogenesis initiates a new cycle of imprinting. Parental genomic imprinting has been detected in plants and relies on DNA methylation by the methyltransferase MET1. However, in contrast to mammals, plant imprints are created by differential removal of silencing marks during gametogenesis. In Arabidopsis, DNA demethylation is mediated by the DNA glycosylase DEMETER (DME) causing activation of imprinted genes at the end of female gametogenesis. On the basis of genetic interactions, we show that in addition to DME, the plant homologs of the human Retinoblastoma (Rb) and its binding partner RbAp48 are required for the activation of the imprinted genes FIS2 and FWA. This Rb-dependent activation is mediated by direct transcriptional repression of MET1 during female gametogenesis. We have thus identified a new mechanism required for imprinting establishment, outlining a new role for the Retinoblastoma pathway, which may be conserved in mammals.
Author Summary
Imprinting in plants and mammals involves a process whereby one of the two inherited gene variants (alleles) is inactivated. During imprinting, the transcriptional silencing of one allele is mediated by histone modifications or DNA methylation. The expressed parental allele is activated during gametogenesis by poorly understood mechanisms that remove silencing marks. In Arabidopsis, we studied genes expressed only from the maternal allele because the paternal allele is silenced by DNA methylation. We report that the expression of the maternal allele requires the repression of transcription of the major DNA methyltransferase by the sustained activity of the Arabidopsis homologs of the Retinoblastoma pathway. Repression is confined to the female gamete and is essential for the expression of imprinted genes in plants. The conserved transcriptional repression of DNA methyltransferases by the Retinoblastoma pathway suggests that this new regulation of imprinting might be also active in mammals.
Abstract
A new regulation of imprinting discovered in Arabidopsis involves the Retinoblastoma gene.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning.
- Record: found
- Abstract: found
- Article: not found
Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting.
- Record: found
- Abstract: found
- Article: not found
