- Record: found
- Abstract: found
- Article: found
Division of labour between Myc and G1 cyclins in cell cycle commitment and pace control
Read this article at
Abstract
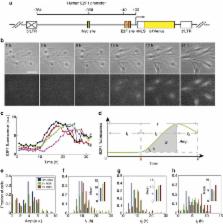
A body of evidence has shown that the control of E2F transcription factor activity is critical for determining cell cycle entry and cell proliferation. However, an understanding of the precise determinants of this control, including the role of other cell-cycle regulatory activities, has not been clearly defined. Here, recognizing that the contributions of individual regulatory components could be masked by heterogeneity in populations of cells, we model the potential roles of individual components together with the use of an integrated system to follow E2F dynamics at the single-cell level and in real time. These analyses reveal that crossing a threshold amplitude of E2F accumulation determines cell cycle commitment. Importantly, we find that Myc is critical in modulating the amplitude, whereas cyclin D/E activities have little effect on amplitude but do contribute to the modulation of duration of E2F activation, thereby affecting the pace of cell cycle progression.
Abstract
 The transcription factor E2F is critical for determining cell proliferation. By monitoring
E2F activity in single cells throughout the cell cycle, Dong
et al. provide evidence that Myc and G1 cyclin/CDKs regulate different aspects of E2F temporal
dynamics, resulting in distinct phenotypic outputs.
The transcription factor E2F is critical for determining cell proliferation. By monitoring
E2F activity in single cells throughout the cell cycle, Dong
et al. provide evidence that Myc and G1 cyclin/CDKs regulate different aspects of E2F temporal
dynamics, resulting in distinct phenotypic outputs.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Encoding and decoding cellular information through signaling dynamics.
- Record: found
- Abstract: found
- Article: not found
p53 dynamics control cell fate.
- Record: found
- Abstract: found
- Article: not found