- Record: found
- Abstract: found
- Article: not found
Using thermodynamic equilibrium models to predict the effect of antiviral agents on infectivity: Theoretical application to SARS-CoV-2 and other viruses.
Read this article at
Abstract
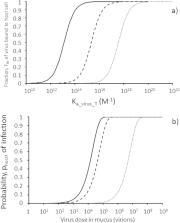
Thermodynamic equilibrium models predict the infectivity of novel and emerging viruses using molecular data including the binding affinity of the virus to the host cell (as represented by the association constant K a_virus_T) and the probability, p virogenesis, of the virus replicating after entry to the cell. Here those models are adapted based on the principles of ligand binding to macromolecules to assess the effect on virus infectivity of inhibitor molecules which target specific proteins of the virus. Three types of inhibitor are considered using the thermodynamic equilibrium model for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection of the human lung with parameters for the strength and nature of the interaction between the target virus protein and the inhibitor molecule. The first is competitive inhibition of the SARS-CoV-2 spike glycoprotein (SGP) trimer binding to its human angiotensin converting enzyme 2 (ACE2) receptor by unfractionated heparin (UFH). Using a novel approach presented here, a value of K a_virus_T = 3.53 × 10 17 M −1 is calculated for SARS-CoV-2 from the IC 50 for inhibition by UFH of SARS-CoV-2 plaque formation in cell culture together with the dissociation constant K VI of 0.73 × 10 −10 M reported for heparin binding to SARS-CoV-2 SGP trimer. Such a high K a_virus_T limits the effectiveness of a competitive inhibitors such as UFH. The second is the attachment of a nanoparticle such as a zinc oxide tetrapod (ZnOT) to the virus shell as for herpes simplex virus (HSV). The increase in molecular weight through ZnOT attachment is predicted to decrease K a_virus_T by orders of magnitude by making the entropy change (ΔS a_immob) on immobilisation of the ZnOT:virus complex on cell binding more negative than for the virus alone. According to the model, ZnOT acts synergistically with UFH at the IC 50 of 33 μg/cm 3 which together decrease viral infectivity by 61,000-fold compared to the two-fold and three-fold decreases predicted for UFH alone at the IC 50 and for ZnOT alone respectively. According to the model here, UFH alone at its peak deliverable dose to the lung of 1,000 μg/cm 3 only decreases infectivity by 31-fold. Practicable approaches to target and decrease ΔS a_immob for respiratory viruses should therefore be considered. The combination of decreasing ΔS a_immob together with blocking the interaction of virus surface protein with its host cell receptor may achieve synergistic effects for faecal-oral viruses and HSV. The third is reversible noncompetitive inhibition of the viral main protease (M pro) for which the decrease in p virogenesis is assumed to be proportional to the decrease in enzyme activity as predicted by enzyme kinetic equations for a given concentration of inhibitor which binds to M pro with dissociation constant K i. Virologists reporting viral inhibition studies are urged to report the concentration of cells in the cell culture experiment as this is a key parameter in estimating K a_virus_T here.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: found
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
- Record: found
- Abstract: found
- Article: found
Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors
- Record: found
- Abstract: found
- Article: not found
