- Record: found
- Abstract: found
- Article: found
Methylotrophic Communities Associated with a Greenland Ice Sheet Methane Release Hotspot
Read this article at
Abstract
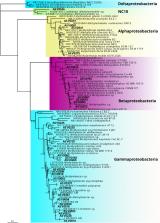
Subglacial environments provide conditions suitable for the microbial production of methane, an important greenhouse gas, which can be released from beneath the ice as a result of glacial melting. High gaseous methane emissions have recently been discovered at Russell Glacier, an outlet of the southwestern margin of the Greenland Ice Sheet, acting not only as a potential climate amplifier but also as a substrate for methane consuming microorganisms. Here, we describe the composition of the microbial assemblage exported in meltwater from the methane release hotspot at Russell Glacier and its changes over the melt season and as it travels downstream. We found that a substantial part (relative abundance 27.2% across the whole dataset) of the exported assemblage was made up of methylotrophs and that the relative abundance of methylotrophs increased as the melt season progressed, likely due to the seasonal development of the glacial drainage system. The methylotrophs were dominated by representatives of type I methanotrophs from the Gammaproteobacteria; however, their relative abundance decreased with increasing distance from the ice margin at the expense of type II methanotrophs and/or methylotrophs from the Alphaproteobacteria and Betaproteobacteria. Our results show that subglacial methane release hotspot sites can be colonized by microorganisms that can potentially reduce methane emissions.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data
- Record: found
- Abstract: not found
- Article: not found
Cutadapt removes adapter sequences from high-throughput sequencing reads
- Record: found
- Abstract: found
- Article: found