- Record: found
- Abstract: found
- Article: found
Stabilization of SIRT7 deacetylase by viral oncoprotein HBx leads to inhibition of growth restrictive RPS7 gene and facilitates cellular transformation
Read this article at
Abstract
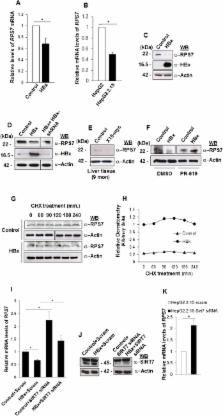
Sirtuin-7 (SIRT7) deacetylase exhibits a high selectivity for acetylated H3K18 and has been implicated in the maintenance of malignant phenotype. However, it remains unclear if SIRT7 and H3K18ac play a role in the tumorigenic program driven by oncogenic viruses. We show that ectopically expressed HBx oncoprotein of hepatitis B virus promoted intracellular stability of SIRT7 by salvaging it from ubiquitin-mediated proteasomal degradation. HBx-dependent accumulation of SIRT7 favored H3K18 deacetylation and down-regulated the small ribosomal protein gene, RPS7, involved in cell death and DNA damage response. HBx facilitated the recruitment of SIRT7 to RPS7 promoter thus impeding H3K18ac occupancy and hindering RPS7 transcription. The antagonistic relationship between SIRT7 and RPS7 was also observed in the HBx transgenic mice, where elevated levels of SIRT7 protein were coincident with low levels of H3K18ac and RPS7. Strikingly, inhibition of cellular deubiquitinase activity restored RPS7 gene transcription. Further, depletion of endogenous SIRT7 led to decreased cell viability and transformation. The biological relevance of RPS7 suppression by HBx-SIRT7 axis was evident from ectopic expression of RPS7 which attenuated clonogenicity of cells. Thus, our findings suggest that SIRT7 is a critical regulator of HBx-driven oncogenic program, through its antagonistic impact on growth restrictive ribosomal protein RPS7.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes.
- Record: found
- Abstract: found
- Article: not found
Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b.
- Record: found
- Abstract: found
- Article: not found