- Record: found
- Abstract: found
- Article: found
Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2
Read this article at
Significance
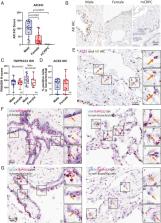
New therapeutic targets are urgently needed against SARS-CoV-2, the coronavirus responsible for the COVID-19 pandemic. Results in this study show that targeting the transcriptional regulation of host entry factors TMPRSS2 and ACE2 is a viable treatment strategy to prevent SARS-CoV-2 infection. In particular, inhibitors of androgen receptor (AR) or bromodomain and extraterminal domain (BET) proteins are effective against SARS-CoV-2 infection. AR inhibitors are already approved in the clinic for treatment of prostate cancer and are under investigation in COVID-19 patients; BET inhibitors are also in clinical development for other indications and could be rapidly repurposed for COVID-19.
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for COVID-19, employs two key host proteins to gain entry and replicate within cells, angiotensin-converting enzyme 2 (ACE2) and the cell surface transmembrane protease serine 2 (TMPRSS2). TMPRSS2 was first characterized as an androgen-regulated gene in the prostate. Supporting a role for sex hormones, males relative to females are disproportionately affected by COVID-19 in terms of mortality and morbidity. Several studies, including one employing a large epidemiological cohort, suggested that blocking androgen signaling is protective against COVID-19. Here, we demonstrate that androgens regulate the expression of ACE2, TMPRSS2, and androgen receptor (AR) in subsets of lung epithelial cells. AR levels are markedly elevated in males relative to females greater than 70 y of age. In males greater than 70 y old, smoking was associated with elevated levels of AR and ACE2 in lung epithelial cells. Transcriptional repression of the AR enhanceosome with AR or bromodomain and extraterminal domain (BET) antagonists inhibited SARS-CoV-2 infection in vitro. Taken together, these studies support further investigation of transcriptional inhibition of critical host factors in the treatment or prevention of COVID-19.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor
- Record: found
- Abstract: found
- Article: not found
Comprehensive Integration of Single-Cell Data

- Record: found
- Abstract: found
- Article: found