- Record: found
- Abstract: found
- Article: found
Rapid expansion and long-term persistence of elevated NK cell numbers in humans infected with hantavirus
Read this article at
Abstract
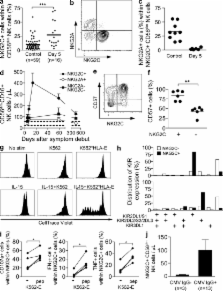
Acute hantavirus infection in humans triggers a rapid expansion and long-term persistence of NK cells.
Abstract
Natural killer (NK) cells are known to mount a rapid response to several virus infections. In experimental models of acute viral infection, this response has been characterized by prompt NK cell activation and expansion followed by rapid contraction. In contrast to experimental model systems, much less is known about NK cell responses to acute viral infections in humans. We demonstrate that NK cells can rapidly expand and persist at highly elevated levels for >60 d after human hantavirus infection. A large part of the expanding NK cells expressed the activating receptor NKG2C and were functional in terms of expressing a licensing inhibitory killer cell immunoglobulin-like receptor (KIR) and ability to respond to target cell stimulation. These results demonstrate that NK cells can expand and remain elevated in numbers for a prolonged period of time in humans after a virus infection. In time, this response extends far beyond what is considered normal for an innate immune response.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C.
- Record: found
- Abstract: found
- Article: not found
Immunology of hepatitis B virus and hepatitis C virus infection.
- Record: found
- Abstract: found
- Article: not found