- Record: found
- Abstract: found
- Article: not found
Expression of HDAC2 but Not HDAC1 Transcript Is Reduced in Dorsolateral Prefrontal Cortex of Patients with Schizophrenia
Read this article at
Abstract

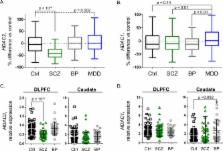
Postmortem brain studies support dysregulated expression of the histone deacetylase enzymes, HDAC1 and HDAC2, as a central feature in diseases including schizophrenia, bipolar disorder, and depression. Our objective was to investigate HDAC expression in a large postmortem sample set representing healthy and disease brains. We used >700 well-characterized samples from patients diagnosed with schizophrenia ( n = 175), major depressive disorder ( n = 135), and bipolar disorder ( n = 61) to measure HDAC1 and HDAC2 transcript levels by quantitative real-time PCR in dorsolateral prefrontal cortex (DLPFC) and caudate compared to control samples. HDAC expression was calculated relative to the geometric mean of β-2-microglobulin, β-glucuronidase, and β-actin. In adult-age DLPFC, HDAC2 was decreased by 34% in schizophrenia samples compared to controls ( p < 10 –4). HDAC2 was significantly upregulated in major depressive disorder samples by 17% versus controls ( p = 0.002). Neither smoking history nor therapeutic drugs impacted HDAC2 levels and no HDAC1 patient-control differences were observed. In caudate, HDAC levels were unchanged between patient and control groups. In control DLPFC, age fetal week 14 to 97 years ( n = 326), both HDAC1 and HDAC2 levels sharply declined around birth and stabilized thereafter. Using by far the largest postmortem sample set on this topic, our major finding (decreased HDAC2 transcript) showed notable specificity in disease (schizophrenia but not major depressive disorder), HDAC subtype ( HDAC2 but not HDAC1) and brain region (DLPFC but not caudate). These differences shape understanding of regional components of neural circuitry in the diseased brain and set a benchmark to quantify HDAC density and distribution using in vivo neuroimaging tools.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Temporal dynamics and genetic control of transcription in the human prefrontal cortex.
- Record: found
- Abstract: found
- Article: not found
Antidepressant actions of histone deacetylase inhibitors.
- Record: found
- Abstract: found
- Article: not found
