- Record: found
- Abstract: found
- Article: found
PKC Signaling Regulates Drug Resistance of the Fungal Pathogen Candida albicans via Circuitry Comprised of Mkc1, Calcineurin, and Hsp90
Read this article at
Abstract
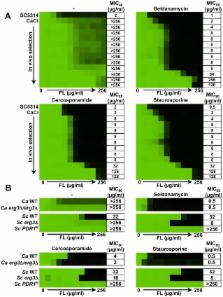
Fungal pathogens exploit diverse mechanisms to survive exposure to antifungal drugs. This poses concern given the limited number of clinically useful antifungals and the growing population of immunocompromised individuals vulnerable to life-threatening fungal infection. To identify molecules that abrogate resistance to the most widely deployed class of antifungals, the azoles, we conducted a screen of 1,280 pharmacologically active compounds. Three out of seven hits that abolished azole resistance of a resistant mutant of the model yeast Saccharomyces cerevisiae and a clinical isolate of the leading human fungal pathogen Candida albicans were inhibitors of protein kinase C (PKC), which regulates cell wall integrity during growth, morphogenesis, and response to cell wall stress. Pharmacological or genetic impairment of Pkc1 conferred hypersensitivity to multiple drugs that target synthesis of the key cell membrane sterol ergosterol, including azoles, allylamines, and morpholines. Pkc1 enabled survival of cell membrane stress at least in part via the mitogen activated protein kinase (MAPK) cascade in both species, though through distinct downstream effectors. Strikingly, inhibition of Pkc1 phenocopied inhibition of the molecular chaperone Hsp90 or its client protein calcineurin. PKC signaling was required for calcineurin activation in response to drug exposure in S. cerevisiae. In contrast, Pkc1 and calcineurin independently regulate drug resistance via a common target in C. albicans. We identified an additional level of regulatory control in the C. albicans circuitry linking PKC signaling, Hsp90, and calcineurin as genetic reduction of Hsp90 led to depletion of the terminal MAPK, Mkc1. Deletion of C. albicans PKC1 rendered fungistatic ergosterol biosynthesis inhibitors fungicidal and attenuated virulence in a murine model of systemic candidiasis. This work establishes a new role for PKC signaling in drug resistance, novel circuitry through which Hsp90 regulates drug resistance, and that targeting stress response signaling provides a promising strategy for treating life-threatening fungal infections.
Author Summary
Treating fungal infections is challenging due to the emergence of drug resistance and the limited number of clinically useful antifungal drugs. We screened a library of 1,280 pharmacologically active compounds to identify those that reverse resistance of the leading human fungal pathogen, Candida albicans, to the most widely used antifungals, the azoles. This revealed a new role for protein kinase C (PKC) signaling in resistance to drugs targeting the cell membrane, including azoles, allylamines, and morpholines. We dissected mechanisms through which PKC regulates resistance in C. albicans and the model yeast Saccharomyces cerevisiae. PKC enabled survival of cell membrane stress at least in part through the mitogen-activated protein kinase (MAPK) cascade in both species. In S. cerevisiae, inhibition of PKC signaling blocked activation of a key regulator of membrane stress responses, calcineurin. In C. albicans, Pkc1 and calcineurin independently regulate resistance via a common target. Deletion of C. albicans PKC1 rendered fungistatic drugs fungicidal and reduced virulence in a mouse model. The molecular chaperone Hsp90, which stabilizes client proteins including calcineurin, also stabilized the terminal C. albicans MAPK, Mkc1. We establish new circuitry connecting PKC with Hsp90 and calcineurin and suggest a promising strategy for treating life-threatening fungal infections.
Related collections
Most cited references70
- Record: found
- Abstract: found
- Article: not found
Fungal secondary metabolism - from biochemistry to genomics.
- Record: found
- Abstract: found
- Article: not found
Cell wall integrity signaling in Saccharomyces cerevisiae.
- Record: found
- Abstract: found
- Article: not found