- Record: found
- Abstract: found
- Article: found
Fungal-Bacterial Interactions in Health and Disease
review-article

Wibke Krüger
1 ,
Sarah Vielreicher
1 ,
Mario Kapitan
1
,
2 ,
Ilse D. Jacobsen
1
,
2
,
3 ,
Maria Joanna Niemiec
1
,
2
,
*
21 May 2019
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
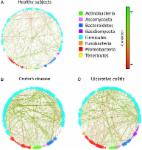
Fungi and bacteria encounter each other in various niches of the human body. There, they interact directly with one another or indirectly via the host response. In both cases, interactions can affect host health and disease. In the present review, we summarized current knowledge on fungal-bacterial interactions during their commensal and pathogenic lifestyle. We focus on distinct mucosal niches: the oral cavity, lung, gut, and vagina. In addition, we describe interactions during bloodstream and wound infections and the possible consequences for the human host.
Related collections
Most cited references287
- Record: found
- Abstract: found
- Article: found
Fungal microbiota dysbiosis in IBD
Harry Sokol, Valentin Leducq, Hugues Aschard … (2016)
- Record: found
- Abstract: found
- Article: found
Defining the healthy "core microbiome" of oral microbial communities
Egija Zaura, Bart JF Keijser, Susan M. Huse … (2009)
- Record: found
- Abstract: found
- Article: not found
Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis.
Iliyan Iliev, Vincent Funari, Kent D. Taylor … (2012)