- Record: found
- Abstract: found
- Article: not found
Murine Leukemias with Retroviral Insertions at Lmo2 Are Predictive of the Leukemias Induced in SCID-X1 Patients Following Retroviral Gene Therapy
Read this article at
Abstract
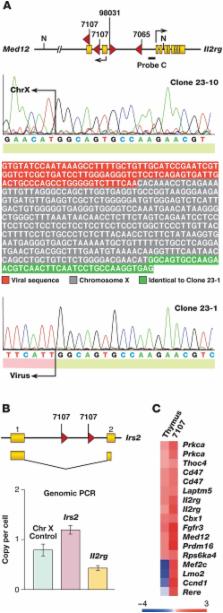
Five X-linked severe combined immunodeficiency patients (SCID-X1) successfully treated with autologous bone marrow stem cells infected ex vivo with an IL2RG-containing retrovirus subsequently developed T-cell leukemia and four contained insertional mutations at LMO2. Genetic evidence also suggests a role for IL2RG in tumor formation, although this remains controversial. Here, we show that the genes and signaling pathways deregulated in murine leukemias with retroviral insertions at Lmo2 are similar to those deregulated in human leukemias with high LMO2 expression and are highly predictive of the leukemias induced in SCID-X1 patients. We also provide additional evidence supporting the notion that IL2RG and LMO2 cooperate in leukemia induction but are not sufficient and require additional cooperating mutations. The highly concordant nature of the genetic events giving rise to mouse and human leukemias with mutations at Lmo2 are an encouraging sign to those wanting to use mice to model human cancer and may help in designing safer methods for retroviral gene therapy.
Author Summary
Twenty patients with X-linked severe combined immunodeficiency (SCID-X1) have been successfully treated by gene therapy. Unfortunately, five of these patients have developed T-cell leukemia two or more years after receiving the therapeutic gene IL2RG on a retroviral vector. The leukemias developed because the vector inserted itself near cancer-causing genes and disrupted their normal regulation. Remarkably, in four patients, the vector inserted near a known T-cell oncogene, LMO2. We have found that in mice, similar retroviruses cause T-cell leukemias by inserting near Lmo2. We have found two leukemias that have retroviral insertions near Lmo2 and Il2rg in the same cell. The probability of these insertions happening by chance is exceedingly small and these results imply that these two genes are deregulated together to induce leukemia. Our data show that Lmo2 and Il2rg cooperate but may not be sufficient for leukemia development and additional mutations contribute to leukemia development. We have also found cooperating retroviral insertions in genes that are abnormally expressed in human T-cell leukemias. The mouse models provide unique insight into the pathogenesis of T-cell leukemia, and they are highly predictive of the leukemias caused by SCID-X1 gene therapy.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1.
- Record: found
- Abstract: found
- Article: not found
Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling.
- Record: found
- Abstract: found
- Article: not found
