- Record: found
- Abstract: found
- Article: found
Constitutive overexpression of the TaNF-YB4 gene in transgenic wheat significantly improves grain yield
Read this article at
Highlight
Constitutive overexpression of NF-YB in transgenic wheat leads to a 20–30% increase in grain yield compared with wild-type plants (cv. Gladius) without negative changes in seed size or number.
Abstract
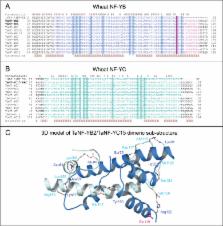
Heterotrimeric nuclear factors Y (NF-Ys) are involved in regulation of various vital functions in all eukaryotic organisms. Although a number of NF-Y subunits have been characterized in model plants, only a few have been functionally evaluated in crops. In this work, a number of genes encoding NF-YB and NF-YC subunits were isolated from drought-tolerant wheat ( Triticum aestivum L. cv. RAC875), and the impact of the overexpression of TaNF-YB4 in the Australian wheat cultivar Gladius was investigated. TaNF-YB4 was isolated as a result of two consecutive yeast two-hybrid (Y2H) screens, where ZmNF-YB2a was used as a starting bait. A new NF-YC subunit, designated TaNF-YC15, was isolated in the first Y2H screen and used as bait in a second screen, which identified two wheat NF-YB subunits, TaNF-YB2 and TaNF-YB4. Three-dimensional modelling of a TaNF-YB2/TaNF-YC15 dimer revealed structural determinants that may underlie interaction selectivity. The TaNF-YB4 gene was placed under the control of the strong constitutive polyubiquitin promoter from maize and introduced into wheat by biolistic bombardment. The growth and yield components of several independent transgenic lines with up-regulated levels of TaNF-YB4 were evaluated under well-watered conditions (T 1–T 3 generations) and under mild drought (T 2 generation). Analysis of T 2 plants was performed in large deep containers in conditions close to field trials. Under optimal watering conditions, transgenic wheat plants produced significantly more spikes but other yield components did not change. This resulted in a 20–30% increased grain yield compared with untransformed control plants. Under water-limited conditions transgenic lines maintained parity in yield performance.
Related collections
Most cited references55
- Record: found
- Abstract: not found
- Article: not found
Identification of common molecular subsequences.
- Record: found
- Abstract: not found
- Article: not found
TreeView: an application to display phylogenetic trees on personal computers.

- Record: found
- Abstract: found
- Article: found