- Record: found
- Abstract: found
- Article: found
Brain age and other bodily ‘ages’: implications for neuropsychiatry
Read this article at
Abstract
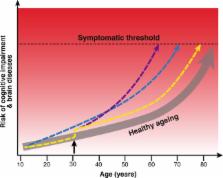
As our brains age, we tend to experience cognitive decline and are at greater risk of neurodegenerative disease and dementia. Symptoms of chronic neuropsychiatric diseases are also exacerbated during ageing. However, the ageing process does not affect people uniformly; nor, in fact, does the ageing process appear to be uniform even within an individual. Here, we outline recent neuroimaging research into brain ageing and the use of other bodily ageing biomarkers, including telomere length, the epigenetic clock, and grip strength. Some of these techniques, using statistical approaches, have the ability to predict chronological age in healthy people. Moreover, they are now being applied to neurological and psychiatric disease groups to provide insights into how these diseases interact with the ageing process and to deliver individualised predictions about future brain and body health. We discuss the importance of integrating different types of biological measurements, from both the brain and the rest of the body, to build more comprehensive models of the biological ageing process. Finally, we propose seven steps for the field of brain-ageing research to take in coming years. This will help us reach the long-term goal of developing clinically applicable statistical models of biological processes to measure, track and predict brain and body health in ageing and disease.
Related collections
Most cited references84
- Record: found
- Abstract: found
- Article: not found
Oxidative stress shortens telomeres.

- Record: found
- Abstract: found
- Article: found
DNA methylation age of blood predicts all-cause mortality in later life
- Record: found
- Abstract: found
- Article: not found