- Record: found
- Abstract: found
- Article: found
A potent neutralizing nanobody against SARS‐CoV‐2 with inhaled delivery potential

Read this article at
Abstract
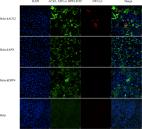
The coronavirus disease 2019 (COVID‐19) pandemic has become a serious burden on global public health. Although therapeutic drugs against COVID‐19 have been used in many countries, their efficacy is still limited. We here reported nanobody (Nb) phage display libraries derived from four camels immunized with the SARS‐CoV‐2 spike receptor‐binding domain (RBD), from which 381 Nbs were identified to recognize SARS‐CoV‐2‐RBD. Furthermore, seven Nbs were shown to block interaction of human angiotensin‐converting enzyme 2 (ACE2) with SARS‐CoV‐2‐RBD variants and two Nbs blocked the interaction of human ACE2 with bat‐SL‐CoV‐WIV1‐RBD and SARS‐CoV‐1‐RBD. Among these candidates, Nb11‐59 exhibited the highest activity against authentic SARS‐CoV‐2 with 50% neutralizing dose (ND 50) of 0.55 μg/ml. Nb11‐59 can be produced on large scale in Pichia pastoris, with 20 g/L titer and 99.36% purity. It also showed good stability profile, and nebulization did not impact its stability. Overall, Nb11‐59 might be a promising prophylactic and therapeutic molecule against COVID‐19, especially through inhalation delivery.
Abstract
We reported nanobody (Nb) phage display libraries derived from four camels immunized with the SARS‐CoV‐2 spike receptor‐binding domain (RBD), from which 381 Nbs were identified to recognize SARS‐CoV‐2‐RBD including several mutants. Nb11‐59 exhibited potent antiviral activity against authentic SARS‐CoV‐2 with 50% neutralizing dose (ND 50) of 0.55 μg/ml, and it can be produced on large scale in Pichia pastoris with titers reached 20 g/L. Importantly, Nb11‐59 showed a good stability and could be developed as an inhaled drug to treat COVID‐19.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found
Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor

- Record: found
- Abstract: found
- Article: found
