- Record: found
- Abstract: found
- Article: found
Gut microbiome composition, not alpha diversity, is associated with survival in a natural vertebrate population
Read this article at
Abstract
Background
The vertebrate gut microbiome (GM) can vary substantially across individuals within the same natural population. Although there is evidence linking the GM to health in captive animals, very little is known about the consequences of GM variation for host fitness in the wild. Here, we explore the relationship between faecal microbiome diversity, body condition, and survival using data from the long-term study of a discrete natural population of the Seychelles warbler ( Acrocephalus sechellensis) on Cousin Island. To our knowledge, this is the first time that GM differences associated with survival have been fully characterised for a natural vertebrate species, across multiple age groups and breeding seasons.
Results
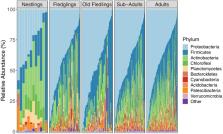
We identified substantial variation in GM community structure among sampled individuals, which was partially explained by breeding season (5% of the variance), and host age class (up to 1% of the variance). We also identified significant differences in GM community membership between adult birds that survived, versus those that had died by the following breeding season. Individuals that died carried increased abundances of taxa that are known to be opportunistic pathogens, including several ASVs in the genus Mycobacterium. However, there was no association between GM alpha diversity (the diversity of bacterial taxa within a sample) and survival to the next breeding season, or with individual body condition. Additionally, we found no association between GM community membership and individual body condition.
Conclusions
These results demonstrate that components of the vertebrate GM can be associated with host fitness in the wild. However, further research is needed to establish whether changes in bacterial abundance contribute to, or are only correlated with, differential survival; this will add to our understanding of the importance of the GM in the evolution of host species living in natural populations.
Related collections
Most cited references116
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: not found
- Article: not found
Fitting Linear Mixed-Effects Models Usinglme4
- Record: found
- Abstract: found
- Article: not found