- Record: found
- Abstract: found
- Article: not found
Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing
Read this article at
Abstract
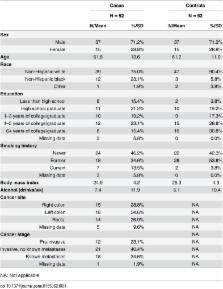
Accumulating evidence indicates that the gut microbiota affects colorectal cancer development, but previous studies have varied in population, technical methods, and associations with cancer. Understanding these variations is needed for comparisons and for potential pooling across studies. Therefore, we performed whole-genome shotgun sequencing on fecal samples from 52 pre-treatment colorectal cancer cases and 52 matched controls from Washington, DC. We compared findings from a previously published 16S rRNA study to the metagenomics-derived taxonomy within the same population. In addition, metagenome-predicted genes, modules, and pathways in the Washington, DC cases and controls were compared to cases and controls recruited in France whose specimens were processed using the same platform. Associations between the presence of fecal Fusobacteria, Fusobacterium, and Porphyromonas with colorectal cancer detected by 16S rRNA were reproduced by metagenomics, whereas higher relative abundance of Clostridia in cancer cases based on 16S rRNA was merely borderline based on metagenomics. This demonstrated that within the same sample set, most, but not all taxonomic associations were seen with both methods. Considering significant cancer associations with the relative abundance of genes, modules, and pathways in a recently published French metagenomics dataset, statistically significant associations in the Washington, DC population were detected for four out of 10 genes, three out of nine modules, and seven out of 17 pathways. In total, colorectal cancer status in the Washington, DC study was associated with 39% of the metagenome-predicted genes, modules, and pathways identified in the French study. More within and between population comparisons are needed to identify sources of variation and disease associations that can be reproduced despite these variations. Future studies should have larger sample sizes or pool data across studies to have sufficient power to detect associations that are reproducible and significant after correction for multiple testing.
Related collections
Most cited references11
- Record: found
- Abstract: found
- Article: not found
Relating the metatranscriptome and metagenome of the human gut.
- Record: found
- Abstract: found
- Article: not found
Accurate and universal delineation of prokaryotic species.
- Record: found
- Abstract: found
- Article: not found
