- Record: found
- Abstract: found
- Article: found
Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome

Read this article at
Summary
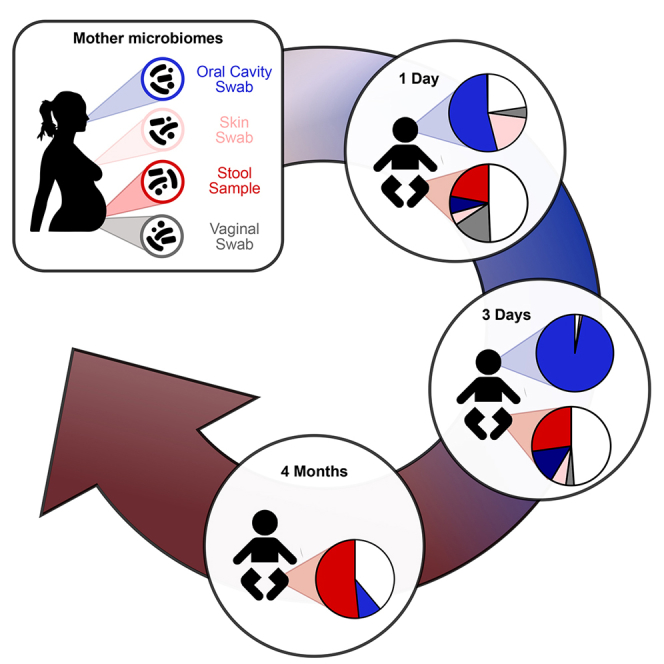
The acquisition and development of the infant microbiome are key to establishing a healthy host-microbiome symbiosis. The maternal microbial reservoir is thought to play a crucial role in this process. However, the source and transmission routes of the infant pioneering microbes are poorly understood. To address this, we longitudinally sampled the microbiome of 25 mother-infant pairs across multiple body sites from birth up to 4 months postpartum. Strain-level metagenomic profiling showed a rapid influx of microbes at birth followed by strong selection during the first few days of life. Maternal skin and vaginal strains colonize only transiently, and the infant continues to acquire microbes from distinct maternal sources after birth. Maternal gut strains proved more persistent in the infant gut and ecologically better adapted than those acquired from other sources. Together, these data describe the mother-to-infant microbiome transmission routes that are integral in the development of the infant microbiome.
Graphical Abstract
Highlights
-
•
Strain-resolved metagenomics was used to track mother-to-infant microbiome transfer
-
•
Microbial strains from multiple maternal body sites transfer to the infant microbiome
-
•
The early microbial diversity in the infant gut is rapidly shaped by niche selection
-
•
The maternal gut microbiome is the source of the majority of transmitted strains
Abstract
Ferretti et al. use metagenomics with strain-resolved computational profiling to characterize the transfer of microbes from mothers to their infants during their first 4 months of life. Multiple maternal body sites contribute to the developing infant microbiome, with maternal gut strains providing the largest contribution of colonizing microorganisms.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data.
- Record: found
- Abstract: found
- Article: not found
The microbiome and innate immunity.
- Record: found
- Abstract: found
- Article: not found