- Record: found
- Abstract: found
- Article: not found
Phytomedicines Explored under In vitro and In silico Studies against Coronavirus: An Opportunity to Develop Traditional Medicines
Read this article at
Abstract
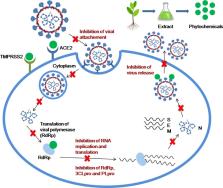
The widespread COVID-19 pandemic, caused by novel coronavirus SARS-CoV-2, has emanated as one of the most life-threatening transmissible diseases. Currently, the repurposed drugs such as remdesivir, azithromycine, chloroquine, and hydroxychloroquine are being employed in the management of COVID-19 but their adverse effects are a matter of concern. In this regard, alternative treatment options i.e., traditional medicine, medicinal plants, and their phytochemicals, which exhibit significant therapeutic efficacy and show a low toxicity profile, are being explored. The current review aims at unraveling the promising medicinal plants, phytochemicals, and traditional medicines against SARS-CoV-2 to discover phytomedicines for the management of COVID-19 on the basis of their potent antiviral activities against coronaviruses, as demonstrated in various biochemical and computational chemical biology studies. The review consists of integrative and updated information on the potential traditional medicines against COVID-19 and will facilitate researchers to develop traditional medicines for the management of COVID-19.
Related collections
Most cited references95
- Record: found
- Abstract: found
- Article: not found
A Novel Coronavirus from Patients with Pneumonia in China, 2019
- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found
