- Record: found
- Abstract: found
- Article: found
Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm
Read this article at
Abstract
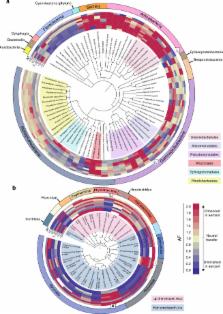
Ocean-derived, airborne microbes play important roles in Earth’s climate system and human health, yet little is known about factors controlling their transfer from the ocean to the atmosphere. Here, we study microbiomes of isolated sea spray aerosol (SSA) collected in a unique ocean–atmosphere facility and demonstrate taxon-specific aerosolization of bacteria and viruses. These trends are conserved within taxonomic orders and classes, and temporal variation in aerosolization is similarly shared by related taxa. We observe enhanced transfer into SSA of Actinobacteria, certain Gammaproteobacteria, and lipid-enveloped viruses; conversely, Flavobacteriia, some Alphaproteobacteria, and Caudovirales are generally under-represented in SSA. Viruses do not transfer to SSA as efficiently as bacteria. The enrichment of mycolic acid-coated Corynebacteriales and lipid-enveloped viruses (inferred from genomic comparisons) suggests that hydrophobic properties increase transport to the sea surface and SSA. Our results identify taxa relevant to atmospheric processes and a framework to further elucidate aerosolization mechanisms influencing microbial and viral transport pathways.
Abstract
Factors controlling the transfer of microbes from the ocean to the atmosphere are unclear. Here, Michaud et al. study this process in an enclosed ocean-atmosphere facility, and show that the degree of aerosolization of bacteria and viruses is taxon-specific.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
The role of particle size in aerosolised pathogen transmission: A review

- Record: found
- Abstract: found
- Article: found
Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage
- Record: found
- Abstract: not found
- Article: not found