- Record: found
- Abstract: found
- Article: found
A chromosome-level reference genome of Ensete glaucum gives insight into diversity and chromosomal and repetitive sequence evolution in the Musaceae
Read this article at
Abstract
Background
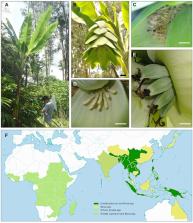
Ensete glaucum (2 n = 2 x = 18) is a giant herbaceous monocotyledonous plant in the small Musaceae family along with banana ( Musa). A high-quality reference genome sequence assembly of E. glaucum is a resource for functional and evolutionary studies of Ensete, Musaceae, and the Zingiberales.
Findings
Using Oxford Nanopore Technologies, chromosome conformation capture (Hi-C), Illumina and RNA survey sequence, supported by molecular cytogenetics, we report a high-quality 481.5 Mb genome assembly with 9 pseudo-chromosomes and 36,836 genes. A total of 55% of the genome is composed of repetitive sequences with predominantly LTR-retroelements (37%) and DNA transposons (7%). The single 5S ribosomal DNA locus had an exceptionally long monomer length of 1,056 bp, more than twice that of the monomers at multiple loci in Musa. A tandemly repeated satellite (1.1% of the genome, with no similar sequence in Musa) was present around all centromeres, together with a few copies of a long interspersed nuclear element (LINE) retroelement. The assembly enabled us to characterize in detail the chromosomal rearrangements occurring between E. glaucum and the x = 11 species of Musa. One E. glaucum chromosome has the same gene content as Musa acuminata, while others show multiple, complex, but clearly defined evolutionary rearrangements in the change between x= 9 and 11.
Conclusions
The advance towards a Musaceae pangenome including E. glaucum, tolerant of extreme environments, makes a complete set of gene alleles, copy number variation, and a reference for structural variation available for crop breeding and understanding environmental responses. The chromosome-scale genome assembly shows the nature of chromosomal fusion and translocation events during speciation, and features of rapid repetitive DNA change in terms of copy number, sequence, and genomic location, critical to understanding its role in diversity and evolution.
Related collections
Most cited references121

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found