- Record: found
- Abstract: found
- Article: found
VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition
Read this article at
Abstract
Background
Taxonomic profiles of vaginal microbial communities can be sorted into a discrete number of categories termed community state types (CSTs). This approach is advantageous because collapsing a hyper-dimensional taxonomic profile into a single categorical variable enables efforts such as data exploration, epidemiological studies, and statistical modeling. Vaginal communities are typically assigned to CSTs based on the results of hierarchical clustering of the pairwise distances between samples. However, this approach is problematic because it complicates between-study comparisons and because the results are entirely dependent on the particular set of samples that were analyzed. We sought to standardize and advance the assignment of samples to CSTs.
Results
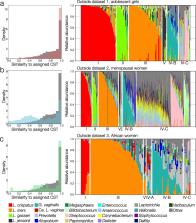
We developed VALENCIA ( VAgina L community state typ E Nearest Centro Id cl Assifier), a nearest centroid-based tool which classifies samples based on their similarity to a set of reference centroids. The references were defined using a comprehensive set of 13,160 taxonomic profiles from 1975 women in the USA. This large dataset allowed us to comprehensively identify, define, and characterize vaginal CSTs common to reproductive age women and expand upon the CSTs that had been defined in previous studies. We validated the broad applicability of VALENCIA for the classification of vaginal microbial communities by using it to classify three test datasets which included reproductive age eastern and southern African women, adolescent girls, and a racially/ethnically and geographically diverse sample of postmenopausal women. VALENCIA performed well on all three datasets despite the substantial variations in sequencing strategies and bioinformatics pipelines, indicating its broad application to vaginal microbiota. We further describe the relationships between community characteristics (vaginal pH, Nugent score) and participant demographics (race, age) and the CSTs defined by VALENCIA.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data

- Record: found
- Abstract: found
- Article: found
The SILVA ribosomal RNA gene database project: improved data processing and web-based tools
- Record: found
- Abstract: found
- Article: not found