- Record: found
- Abstract: found
- Article: found
Expression of classic cadherins and δ-protocadherins in the developing ferret retina
Read this article at
Abstract
Background
Cadherins are a superfamily of calcium-dependent adhesion molecules that play multiple roles in morphogenesis, including proliferation, migration, differentiation and cell-cell recognition. The subgroups of classic cadherins and δ-protocadherins are involved in processes of neural development, such as neurite outgrowth, pathfinding, target recognition, synaptogenesis as well as synaptic plasticity. We mapped the expression of 7 classic cadherins (CDH4, CDH6, CDH7, CDH8, CDH11, CDH14, CDH20) and 8 δ-protocadherins (PCDH1, PCDH7, PCDH8, PCDH9, PCDH10, PCDH11, PCDH17, PCDH18) at representative stages of retinal development and in the mature retina of the ferret by in situ hybridization.
Results
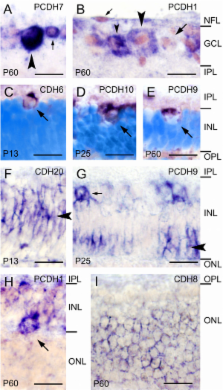
All cadherins investigated by us are expressed differentially by restricted populations of retinal cells during specific periods of the ferret retinogenesis. For example, during embryonic development, some cadherins are exclusively expressed in the outer, proliferative zone of the neuroblast layer, whereas other cadherins mark the prospective ganglion cell layer or cells in the prospective inner nuclear layer. These expression patterns anticipate histogenetic changes that become visible in Nissl or nuclear stainings at later stages. In parallel to the ongoing development of retinal circuits, cadherin expression becomes restricted to specific subpopulations of retinal cell types, especially of ganglion cells, which express most of the investigated cadherins until adulthood. A comparison to previous results in chicken and mouse reveals overall conserved expression patterns of some cadherins but also species differences.
Conclusions
The spatiotemporally restricted expression patterns of 7 classic cadherins and 8 δ-protocadherins indicate that cadherins provide a combinatorial adhesive code that specifies developing retinal cell populations and intraretinal as well as retinofugal neural circuits in the developing ferret retina.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Regulation of cadherin-mediated adhesion in morphogenesis.
- Record: found
- Abstract: found
- Article: not found
Cadherins in development: cell adhesion, sorting, and tissue morphogenesis.
- Record: found
- Abstract: found
- Article: not found