- Record: found
- Abstract: found
- Article: found
Epigenome-wide association study of alcohol use disorder in five brain regions

Read this article at
Abstract
Alcohol use disorder (AUD) is closely linked to the brain regions forming the neurocircuitry of addiction. Postmortem human brain tissue enables the direct study of the molecular pathomechanisms of AUD. This study aims to identify these mechanisms by examining differential DNA-methylation between cases with severe AUD ( n = 53) and controls ( n = 58) using a brain-region-specific approach, in which sample sizes ranged between 46 and 94. Samples of the anterior cingulate cortex (ACC), Brodmann Area 9 (BA9), caudate nucleus (CN), ventral striatum (VS), and putamen (PUT) were investigated. DNA-methylation levels were determined using the Illumina HumanMethylationEPIC Beadchip. Epigenome-wide association analyses were carried out to identify differentially methylated CpG-sites and regions between cases and controls in each brain region. Weighted correlation network analysis (WGCNA), gene-set, and GWAS-enrichment analyses were performed. Two differentially methylated CpG-sites were associated with AUD in the CN, and 18 in VS ( q < 0.05). No epigenome-wide significant CpG-sites were found in BA9, ACC, or PUT. Differentially methylated regions associated with AUD case-/control status ( q < 0.05) were found in the CN ( n = 6), VS ( n = 18), and ACC ( n = 1). In the VS, the WGCNA-module showing the strongest association with AUD was enriched for immune-related pathways. This study is the first to analyze methylation differences between AUD cases and controls in multiple brain regions and consists of the largest sample to date. Several novel CpG-sites and regions implicated in AUD were identified, providing a first basis to explore epigenetic correlates of AUD.
Related collections
Most cited references60
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing

- Record: found
- Abstract: found
- Article: found
WGCNA: an R package for weighted correlation network analysis
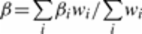
- Record: found
- Abstract: found
- Article: found