- Record: found
- Abstract: found
- Article: found
The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing
Read this article at
Abstract
A common strategy by which bacterial pathogens reside in humans is by shifting from a virulent lifestyle, (systemic infection), to a dormant carrier state. Two major serovars of Salmonella enterica, Typhi and Typhimurium, have evolved a two-component regulatory system to exist inside Salmonella-containing vacuoles in the macrophage, as well as to persist as asymptomatic biofilms in the gallbladder. Here we present evidence that SsrB, a transcriptional regulator encoded on the SPI-2 pathogenicity-island, determines the switch between these two lifestyles by controlling ancestral and horizontally-acquired genes. In the acidic macrophage vacuole, the kinase SsrA phosphorylates SsrB, and SsrB~P relieves silencing of virulence genes and activates their transcription. In the absence of SsrA, unphosphorylated SsrB directs transcription of factors required for biofilm formation specifically by activating csgD ( agfD), the master biofilm regulator by disrupting the silenced, H-NS-bound promoter. Anti-silencing mechanisms thus control the switch between opposing lifestyles.
eLife digest
Salmonella bacteria can infect a range of hosts, including humans and poultry, and cause sickness and diseases such as typhoid fever. Disease-causing Salmonella evolved from harmless bacteria in part by acquiring new genes from other organisms through a process called horizontal gene transfer. However, some strains of disease-causing Salmonella can also survive inside hosts as communities called biofilms without causing any illness to their hosts, who act as carriers of the disease and are able to pass their infection on to others.
So how do Salmonella bacteria ‘decide’ between these two lifestyles? Previous studies have uncovered a regulatory system that controls the decision in Salmonella, which is made up of two proteins called SsrA and SsrB. To trigger the disease-causing lifestyle, SsrA is activated and adds a phosphate group onto SsrB. This in turn causes SsrB to bind to and switch on disease-associated genes in the bacterium. However, it was less clear how the biofilm lifestyle was triggered.
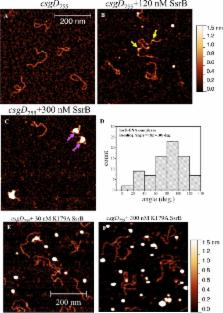
Desai et al. now reveal that the phosphate-free form of SsrB – which was considered to be the inactive form of this protein – plays an important role in the formation of biofilms. Experiments involving an approach called atomic force microscopy showed that the unmodified SsrB acts to stop a major gene that controls biofilm formation from being switched off by a so-called repressor protein.
Salmonella acquired SsrB through horizontal gene transfer, and these findings show how this protein now acts as a molecular switch between disease-causing and biofilm-based lifestyles. SsrB protein is also involved in the decision to switch between these states, but how it does so remains a question for future work.
Related collections
Most cited references45
- Record: found
- Abstract: not found
- Book: not found
Molecular Cloning : A Laboratory Manual
- Record: found
- Abstract: found
- Article: not found
A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa.
- Record: found
- Abstract: found
- Article: not found