- Record: found
- Abstract: found
- Article: found
Multi-omics Comparative Analysis Reveals Multiple Layers of Host Signaling Pathway Regulation by the Gut Microbiota
Read this article at
Abstract
Multiple host pathways were affected by its adaptation to the microbiota. We have found significant transcriptome-proteome discordance caused by the microbiota. This discovery leads to the definite conclusion that transcript-level analysis is not sufficient to predict protein levels and their influence on the function of many specific cellular pathways, so only analysis of combinations of the quantitative data determined at different levels will lead to a complete understanding of the complex relationships between the host and the microbiota. Therefore, our results demonstrate the importance of using an integrative approach to study host-microbiota interaction at the molecular level.
ABSTRACT
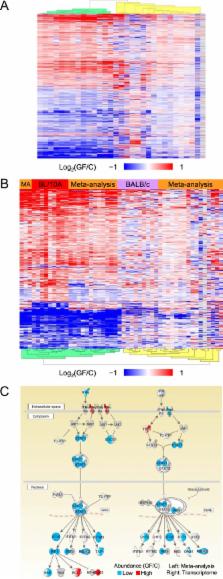
The bodies of mammals are hosts to vast microbial communities composed of trillions of bacteria from thousands of species, whose effects on health and development have begun to be appreciated only recently. In this investigation, an integrated analysis combining proteomics and transcriptomics was used to quantitatively compare the terminal ilia from conventional and germfree mice. Female and male mice responded similarly to the microbiota, but C57BL/10A mice responded more strongly than BALB/c mice at both the transcriptome and proteome levels. The microbiota primarily caused upregulation of immunological pathways and downregulation of metabolic pathways in the conventional mice. Many of the affected pathways were altered only at either the transcriptome or proteome level. Of the pathways that were affected at both levels, most were affected concordantly. The discordant pathways were not principally involved in the immune system but instead were related to metabolism, oxidative phosphorylation, protein translation, transport, and turnover. To broaden the discovery of affected host pathways, a meta-analysis was performed using intestinal transcriptomics data from previously published studies of germfree versus conventional mice with diverse microbiota populations. Similar transcript-level responses to the microbiota were found, and many additional affected host pathways were discovered.
IMPORTANCE Multiple host pathways were affected by its adaptation to the microbiota. We have found significant transcriptome-proteome discordance caused by the microbiota. This discovery leads to the definite conclusion that transcript-level analysis is not sufficient to predict protein levels and their influence on the function of many specific cellular pathways, so only analysis of combinations of the quantitative data determined at different levels will lead to a complete understanding of the complex relationships between the host and the microbiota. Therefore, our results demonstrate the importance of using an integrative approach to study host-microbiota interaction at the molecular level.
Related collections
Most cited references65

- Record: found
- Abstract: found
- Article: found
The Proteomics Identifications (PRIDE) database and associated tools: status in 2013
- Record: found
- Abstract: found
- Article: not found
Activities at the Universal Protein Resource (UniProt)
- Record: found
- Abstract: found
- Article: not found