- Record: found
- Abstract: found
- Article: found
Multigene phylogeny and cell evolution of chromist infrakingdom Rhizaria: contrasting cell organisation of sister phyla Cercozoa and Retaria
Read this article at
Abstract
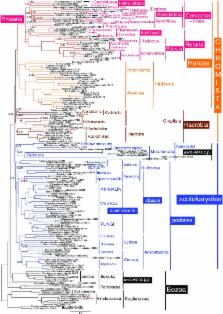
Infrakingdom Rhizaria is one of four major subgroups with distinct cell body plans that comprise eukaryotic kingdom Chromista. Unlike other chromists, Rhizaria are mostly heterotrophic flagellates, amoebae or amoeboflagellates, commonly with reticulose (net-like) or filose (thread-like) feeding pseudopodia; uniquely for eukaryotes, cilia have proximal ciliary transition-zone hub-lattices. They comprise predominantly flagellate phylum Cercozoa and reticulopodial phylum Retaria, whose exact phylogenetic relationship has been uncertain. Given even less clear relationships amongst cercozoan classes, we sequenced partial transcriptomes of seven Cercozoa representing five classes and endomyxan retarian Filoreta marina to establish 187-gene multiprotein phylogenies. Ectoreta (retarian infraphyla Foraminifera, Radiozoa) branch within classical Cercozoa as sister to reticulose Endomyxa. This supports recent transfer of subphylum Endomyxa from Cercozoa to Retaria alongside subphylum Ectoreta which embraces classical retarians where capsules or tests subdivide cells into organelle-containing endoplasm and anastomosing pseudopodial net-like ectoplasm. Cercozoa are more homogeneously filose, often with filose pseudopodia and/or posterior ciliary gliding motility: zooflagellate Helkesimastix and amoeboid Guttulinopsis form a strongly supported clade, order Helkesida. Cercomonads are polyphyletic (Cercomonadida sister to glissomonads; Paracercomonadida deeper). Thecofilosea are a clade, whereas Imbricatea may not be; Sarcomonadea may be paraphyletic. Helkesea and Metromonadea are successively deeper outgroups within cercozoan subphylum Monadofilosa; subphylum Reticulofilosa (paraphyletic on site-heterogeneous trees) branches earliest, Granofilosea before Chlorarachnea. Our multiprotein trees confirm that Rhizaria are sisters of infrakingdom Halvaria (Alveolata, Heterokonta) within chromist subkingdom Harosa (= SAR); they further support holophyly of chromist subkingdom Hacrobia, and are consistent with holophyly of Chromista as sister of kingdom Plantae. Site-heterogeneous rDNA trees group Kraken with environmental DNA clade ‘eSarcomonad’, not Paracercomonadida. Ectoretan fossil dates evidence ultrarapid episodic stem sequence evolution. We discuss early rhizarian cell evolution and multigene tree coevolutionary patterns, gene-paralogue evidence for chromist monophyly, and integrate this with fossil evidence for the age of Rhizaria and eukaryote cells, and revise rhizarian classification.
Related collections
Most cited references189
- Record: found
- Abstract: not found
- Article: not found
Punctuated equilibria: the tempo and mode of evolution reconsidered
- Record: found
- Abstract: found
- Article: not found
Estimating the timing of early eukaryotic diversification with multigene molecular clocks.
- Record: found
- Abstract: found
- Article: not found