- Record: found
- Abstract: not found
- Article: not found
Earth BioGenome Project: Sequencing life for the future of life
Harris A. Lewin ,
Gene E. Robinson ,
W. John Kress ,
William J. Baker ,
Jonathan Coddington ,
Keith A. Crandall ,
Richard Durbin ,
Scott V. Edwards ,
Félix Forest ,
M. Thomas P. Gilbert ,
Melissa M. Goldstein ,
Igor V. Grigoriev ,
Kevin J. Hackett ,
David Haussler ,
Erich D. Jarvis ,
Warren E. Johnson ,
Aristides Patrinos ,
Stephen Richards ,
Juan Carlos Castilla-Rubio ,
Marie-Anne van Sluys ,
Pamela S. Soltis ,
Xun Xu ,
Huanming Yang ,
Guojie Zhang
April 24 2018
April 24 2018
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Increasing our understanding of Earth's biodiversity and responsibly stewarding its
resources are among the most crucial scientific and social challenges of the new millennium.
These challenges require fundamental new knowledge of the organization, evolution,
functions, and interactions among millions of the planet's organisms. Herein, we present
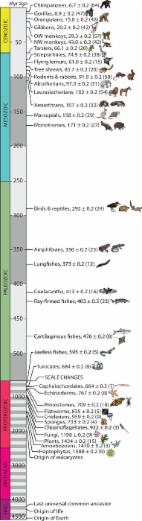
a perspective on the Earth BioGenome Project (EBP), a moonshot for biology that aims
to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity
over a period of 10 years. The outcomes of the EBP will inform a broad range of major
issues facing humanity, such as the impact of climate change on biodiversity, the
conservation of endangered species and ecosystems, and the preservation and enhancement
of ecosystem services. We describe hurdles that the project faces, including data-sharing
policies that ensure a permanent, freely available resource for future scientific
discovery while respecting access and benefit sharing guidelines of the Nagoya Protocol.
We also describe scientific and organizational challenges in executing such an ambitious
project, and the structure proposed to achieve the project's goals. The far-reaching
potential benefits of creating an open digital repository of genomic information for
life on Earth can be realized only by a coordinated international effort.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: found
Tree of Life Reveals Clock-Like Speciation and Diversification
S Blair Hedges, Julie Marin, Michael Suleski … (2015)
- Record: found
- Abstract: found
- Article: not found
Land-use and climate change risks in the Amazon and the need of a novel sustainable development paradigm.
Carlos A. Nobre, Gilvan Sampaio, Laura S Borma … (2016)
- Record: found
- Abstract: found
- Article: not found
Contentious relationships in phylogenomic studies can be driven by a handful of genes
Xing-Xing Shen, Chris Hittinger, Antonis Rokas (2017)
