- Record: found
- Abstract: found
- Article: not found
The Ctenophore Genome and the Evolutionary Origins of Neural Systems
Read this article at
Abstract
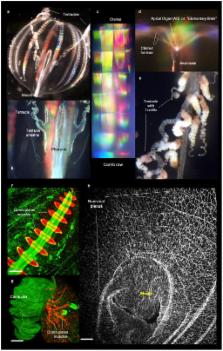
The origins of neural systems remain unresolved. In contrast to other basal metazoans, ctenophores, or comb jellies, have both complex nervous and mesoderm-derived muscular systems. These holoplanktonic predators also have sophisticated ciliated locomotion, behaviour and distinct development. Here, we present the draft genome of Pleurobrachia bachei, Pacific sea gooseberry, together with ten other ctenophore transcriptomes and show that they are remarkably distinct from other animal genomes in their content of neurogenic, immune and developmental genes. Our integrative analyses place Ctenophora as the earliest lineage within Metazoa. This hypothesis is supported by comparative analysis of multiple gene families, including the apparent absence of HOX genes, canonical microRNA machinery, and reduced immune complement in ctenophores. Although two distinct nervous systems are well-recognized in ctenophores, many bilaterian neuron-specific genes and genes of “classical” neurotransmitter pathways either are absent or, if present, are not expressed in neurons. Our metabolomic and physiological data are consistent with the hypothesis that ctenophore neural systems, and possibly muscle specification, evolved independently from those in other animals.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Multiple sequence alignment with the Clustal series of programs.
- Record: found
- Abstract: found
- Article: not found
