- Record: found
- Abstract: found
- Article: found
A shortcut for multiple testing on the directed acyclic graph of gene ontology
Read this article at
Abstract
Background
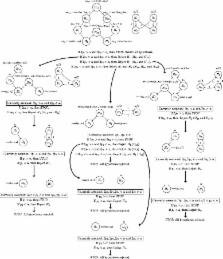
Gene set testing has become an important analysis technique in high throughput microarray and next generation sequencing studies for uncovering patterns of differential expression of various biological processes. Often, the large number of gene sets that are tested simultaneously require some sort of multiplicity correction to account for the multiplicity effect. This work provides a substantial computational improvement to an existing familywise error rate controlling multiplicity approach (the Focus Level method) for gene set testing in high throughput microarray and next generation sequencing studies using Gene Ontology graphs, which we call the Short Focus Level.
Results
The Short Focus Level procedure, which performs a shortcut of the full Focus Level procedure, is achieved by extending the reach of graphical weighted Bonferroni testing to closed testing situations where restricted hypotheses are present, such as in the Gene Ontology graphs. The Short Focus Level multiplicity adjustment can perform the full top-down approach of the original Focus Level procedure, overcoming a significant disadvantage of the otherwise powerful Focus Level multiplicity adjustment. The computational and power differences of the Short Focus Level procedure as compared to the original Focus Level procedure are demonstrated both through simulation and using real data.
Conclusions
The Short Focus Level procedure shows a significant increase in computation speed over the original Focus Level procedure (as much as ∼15,000 times faster). The Short Focus Level should be used in place of the Focus Level procedure whenever the logical assumptions of the Gene Ontology graph structure are appropriate for the study objectives and when either no a priori focus level of interest can be specified or the focus level is selected at a higher level of the graph, where the Focus Level procedure is computationally intractable.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Ontological analysis of gene expression data: current tools, limitations, and open problems.
- Record: found
- Abstract: not found
- Book: not found