- Record: found
- Abstract: found
- Article: not found
A comprehensive methylome map of lineage commitment from hematopoietic progenitors
Read this article at
Abstract
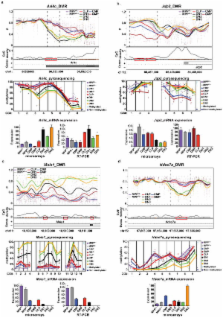
Epigenetic modifications must underlie lineage-specific differentiation as terminally differentiated cells express tissue-specific genes, but their DNA sequence is unchanged. Hematopoiesis provides a well-defined model to study epigenetic modifications during cell-fate decisions, as multipotent progenitors (MPPs) differentiate into progressively restricted myeloid or lymphoid progenitors. While DNA methylation is critical for myeloid versus lymphoid differentiation, as demonstrated by the myeloerythroid bias in Dnmt1 hypomorphs 1, a comprehensive DNA methylation map of hematopoietic progenitors, or of any multipotent/oligopotent lineage, does not exist. Here we examined 4.6 million CpG sites throughout the genome for MPPs, common lymphoid progenitors (CLPs), common myeloid progenitors (CMPs), granulocyte/macrophage progenitors (GMPs), and thymocyte progenitors (DN1, DN2, DN3). Dramatic epigenetic plasticity accompanied both lymphoid and myeloid restriction. Myeloid commitment involved less global DNA methylation than lymphoid commitment, supported functionally by myeloid skewing of progenitors following treatment with a DNA methyltransferase inhibitor. Differential DNA methylation correlated with gene expression more strongly at CpG island shores than CpG islands. Many examples of genes and pathways not previously known to be involved in choice between lymphoid/myeloid differentiation have been identified, such as Arl4c and Jdp2. Several transcription factors, including Meis1, were methylated and silenced during differentiation, suggesting a role in maintaining an undifferentiated state. Additionally, epigenetic modification of modifiers of the epigenome appears to be important in hematopoietic differentiation. Our results directly demonstrate that modulation of DNA methylation occurs during lineage-specific differentiation and defines a comprehensive map of the methylation and transcriptional changes that accompany myeloid versus lymphoid fate decisions.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
CpG islands in vertebrate genomes.
- Record: found
- Abstract: found
- Article: not found
DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45.
- Record: found
- Abstract: found
- Article: not found
