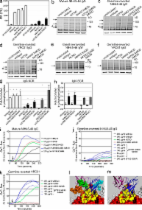
Broadly neutralizing monoclonal antibodies (bnmAbs) that target the structurally conserved CD4 binding site (CD4-BS) bind with high affinity to Env and neutralize diverse HIV-1 isolates, irrespective of their clade (Wu et al., 2010, 2011; Diskin et al., 2011; Scheid et al., 2011). Despite the isolation of these anti–CD4-BS bnmAbs from distinct HIV+ subjects, they share common genetic and structural features, which are critically important for their unique neutralizing properties (Scheid et al., 2011; Wu et al., 2011). So far, four classes of anti–CD4-BS broadly neutralizing antibodies (bNAbs) have been defined: b12, HJ16, VRC01, and 8ANC131 (Kwong and Mascola, 2012). Of particular interest for HIV vaccine development are the VRC01 class antibodies because of their exceptional neutralization potency and breadth. Their VH domains are derived from VH1-2, in contrast to the 8ANC131 class antibodies whose VH domains are derived from VH1-46 (Scheid et al., 2011) and b12 whose VH is derived from VH1-03 (Hoot et al., 2013). The currently known VRC01 class antibodies (such as VRC01, NIH45-46, 3BNC60, and 12A21), were isolated from distinct HIV+ subjects infected with different viruses and display up to 57% diversity in VH and up to 65% diversity in VL sequences; Wu et al., 2010, 2011; Scheid et al., 2011; West et al., 2012; Table S1). Despite this high degree of amino acid sequence diversity, they share structural similarities that allow them to recognize the CD4-BS in a manner very similar to each other and to the CD4 receptor (Scheid et al., 2011; Wu et al., 2011). That interaction is primarily through the VH antibody domains; however, the light chains of VRC01 class antibodies also make important contacts with Env. This contrasts with b12, which appears to interact with Env exclusively through its heavy chain. The mode of Env interaction of HJ16 class antibodies is not yet known. An additional unique feature of the VRC01 class antibodies is that they make contact not only with the gp120 outer domain, but also with the gp120 inner and bridging sheet domains (Diskin et al., 2011; Scheid et al., 2011; Wu et al., 2011). Only a fraction of those infected with HIV develop broadly neutralizing anti–CD4-BS antibodies (Lynch et al., 2012). Despite the presence of anti–CD4-BS epitopes on recombinant Env (Li et al., 2007; Binley et al., 2008; Sather et al., 2009; Mikell et al., 2011), Env immunization has so far failed to elicit such antibodies (Mascola and Montefiori, 2010; Stamatatos, 2012). The reasons why such antibodies are not elicited by Env immunization, and are only rarely generated during natural HIV infection, are currently not well understood. Identifying the roadblocks that prevent the generation of such antibodies and approaches to overcome these barriers will aid in the development of an effective HIV vaccine (Mascola and Montefiori, 2010). The germline-reverted (unmutated) forms of VRC01 class anti–CD4-BS bnmAbs, do not recognize the recombinant Env-derived bait protein used to isolate the B cells expressing the corresponding mature antibodies (Zhou et al., 2010; Scheid et al., 2011). We reported that the germline-reverted forms of two VRC01class antibodies, NIH45-46 (a clonal variant of VRC01; Scheid et al., 2011) and 3BNC60, fail to recognize a large panel of recombinant Env derived from clades A, B, and C, the three most predominant clades worldwide (Hoot et al., 2013). In the current study, we expanded our Env panel and included two additional VRC01 class anti–CD4-BS bnmAbs; 3BNC117 and 12A21 (Scheid et al., 2011; Table S2). The mature versions of these mAbs recognize between 92% (3BNC60) and 71% (12A21) of Envs tested. In contrast, there was no detectable binding to these Envs by the germline-reverted versions of these antibodies. We note that although the accessibility of the CD4-BS by CD4 and anti–CD4-BS antibodies is restricted by the V1, V2, and V3 variable domains of Env and associated N-linked glycosylation sites (NLGS; Wyatt et al., 1993; Ly and Stamatatos, 2000; Saunders et al., 2005), the elimination of these regions did not result in Env-recognition by the germline-reverted anti–CD4-BS bnAbs studied here. The interactions between Env and the germline-reverted forms of anti–CD4-BS bnAbs are commonly investigated using soluble forms (IgG or scFv) of antibodies (Xiao et al., 2009; Wu et al., 2010, 2011; Hoot et al., 2013; Scheid et al., 2011). This interaction may not accurately reflect the interaction of Env with the corresponding membrane-anchored BCRs on B cells. Conceivably, certain recombinant Envs can bind weakly to, and cross-link the germline-reverted BCRs of VRC01 class anti–CD4-BS bNAbs and activate the corresponding B cells. If that is the case, then the deficiency in the development of such antibodies during immunization could not be explained by a lack of recognition of their germline-reverted BCRs by Env. Here, we investigated the interaction between recombinant HIV Env and B cells expressing the germline-reverted BCR versions of VRC01 class antibodies to identify potential roadblocks during the earliest steps of B cell activation. We report that in order for Env to engage and activate B cells expressing germline-reverted NIH45-46 and VRC01 BCRs, specific N-linked glycosylation sites (NLGS) located at key regions of Env must be absent (something that only rarely occurs during natural HIV-infection). RESULTS AND DISCUSSION To ensure that the binding specificities of soluble antibody forms are congruent with their BCR forms, we investigated how diverse Envs interact with B cell–expressed BCRs of mature and germline-reverted NIH45-46; one of the most potent and broadly neutralizing VRC01 class antibodies (Scheid et al., 2011). Similar levels of mature and germline-reverted BCRs were expressed by transfected B cells and both BCRs mediated comparable intracellular Ca2+ flux responses, after cross-linking by anti–human IgG F(ab′)2 (unpublished data). A panel of diverse clade A, B, and C trimeric Envs that display a wide range of affinities for the mature NIH45-46 (mNIH45-46) bound to B cells expressing the mNIH45-46 BCR (Fig. 1, a and b). An Env with a point mutation (D368R), which greatly reduces the binding of anti–CD4-BS antibodies, was included as a negative control. The Envs that bound the mNIH45-46 BCR also induced Ca2+ flux through this BCR (Fig. 1 c). We did not detect binding of these Envs to cells expressing the germline-reverted NIH45-46 BCR (glNIH45-46 BCR; Fig. 1 b), nor did we observe Ca2+ flux in these cells (Fig. 1 d). Similar lack of YU2 Env-binding to the germline-reverted VRC01 (glVRC01) BCR was recently reported (Ota et al., 2012). Figure 1. Env binding to the mature and germline-reverted NIH45-46 BCRs and B cell activation. (a) NIH45-46 IgG binding to the indicated recombinant trimeric gp140 Envs was measured by ELISA. (b) B cells expressing the mature (white bars) or germline-reverted (gray bars) NIH45-46 BCR, as well as mock transfected cells (black bars), were incubated with the indicated trimeric gp140 proteins, followed by incubation with a PE-labeled polyclonal anti-HIV IgG pool. The MFI of PE staining was determined in NIH45-46 BCR expressing cells identified by staining with APC-labeled monoclonal anti–human IgG. Error bars represent SD from the mean of two independent experiments. (c and d) B cells expressing the mature (c) or germline-reverted (d) NIH45-46 BCR were loaded with Fluo-4 direct calcium indicator and stimulated with the indicated trimeric gp140 proteins. Black arrows in c and d indicate time of Env-addition. Data are representative of three independent experiments. Elimination of the Env variable regions 1, 2 or 3 did not result in germline antibody-Env binding (Table S2). However, accessibility of the CD4-BS is also limited by proximal NLGS (Schief et al., 2009). NLGS located in Loop D and V5 of gp120 are of particular interest because they are located in two regions of Env that modulate the neutralizing activities of VRC01 class antibodies (Li et al., 2011). In fact, the light chains of the mature NIH45-46 and VRC01 antibodies contact the N-acetyl-glucosamine residues at Asn 276 in loop D (Zhou et al., 2010; Diskin et al., 2011). We eliminated NLGS from these regions from the clade C Env 426c, which is recognized with high apparent affinity (10.6 nM) by mNIH45-46 (Fig. 1 a, Fig. 2 a, and Table S3). Elimination of the 276, the 460 and 463 (V5), or all three NLGS from 426c increased the binding of mNIH45-46 (apparent affinities of 3.66 nM, 1.9 nM, and 18 pM, respectively; Fig. 2, d, g, and j; and Table S3). The mutant Envs also bound more efficiently to mNIH45-46 BCR (Fig. 3 a) and induced increased B cell activation, as indicated by increases in tyrosine phosphorylation (Fig. 3, b and g) and Ca2+ flux (Fig. 3 i). Importantly, although glNIH45-46 did not bind the 426c Env (Fig. 2 b), it bound the mutant lacking the 276 NLGS (apparent affinity of 0.7 µM; Fig. 2 e and Table S3) and the mutant lacking all three NLGS (apparent affinity of 46 nM; Fig. 2 k and Table S3). Figure 2. Removal of N-linked glycosylation sites from the 426c Env confers binding to germline-reverted NIH45-46 and VRC01. Binding of mature NIH45-46 (a, d, g, and j), germline-reverted NIH45-46 (b, e, h, and k), and germline-reverted VRC01(c, f, i, and l) IgG to trimeric gp140 426c (a–c; wild type), 426c lacking the NLGS at position 276 (d–f; N276D), 426c lacking NLGS at positions 460 and 463 (g–i; N460D.N463D), or 426c lacking NLGS at positions 276, 460, and 463 (j–l; N276D.N460D.N463D) was measured by BLI. For mature and germline-reverted NIH45-46, black, red, orange, blue, and green lines represent 1.7 µM, 850 nM, 425 nM, 212 nM, and 106 nM of trimeric Env, respectively. For germline-reverted VRC01, black, red, orange, blue, and green lines represent 2 µM, 1 µM, 500 nM, 250 nM, and 125 nM of trimeric Env, respectively. Dashed gray lines represent the theoretical fit. The binding kinetic data (including experimental error) are summarized in Table S3. Figure 3. Removal of N-linked glycosylation sites from the 426c Env results in stimulation of the germline-reverted NIH45-46 and VRC01 BCRs. (a) B cells expressing the mature (white bars) or germline-reverted (black bars) NIH45-46 IgG BCR, were incubated with the indicated trimeric gp140 proteins, followed by incubation with a PE-labeled polyclonal anti-HIV IgG pool. The MFI of PE staining was determined in NIH45-46 BCR expressing cells identified by staining with APC-labeled monoclonal anti–human IgG. Error bars represent SD from the mean of two independent experiments (b–f). B cells expressing the mature (b), germline-reverted NIH45-46 (c), and germline-reverted VRC01 (d) IgG BCR, as well as the germline-reverted NIH45-46 (e), and germline-reverted VRC01 (f) IgM BCR were either unstimulated, or were stimulated with the indicated 426c Env variants (40 µg/ml for the mature BCR, or 200 µg/ml for the germline-reverted BCRs). Lysates were separated by SDS-PAGE, transferred to nitrocellulose, and stained with the mouse anti-phosphotyrosine mAb 4G10 (top) and a rabbit anti–β-actin mAb 13E5 (bottom), followed by IRDye 800CW-labeled goat anti–mouse and IRDye 680LT-labeled goat anti–rabbit antibodies. Black arrows indicate proteins that show an increase in tyrosine phosphorylation. (g) Quantification of tyrosine phosphorylation signal corresponding to protein band in b–d are denoted with #. White bars, mature NIH45-46; black bars, germline NIH45-46; gray bars, germline VRC01. (h) Quantification of tyrosine phosphorylation signal corresponding to protein band in e and f, denoted with arrow. Black bars (germline NIH45-46) and gray bars (germline VRC01). Signal intensity was normalized to the actin loading controls, and expressed relative to the unstimulated cells. Error bars represent the SD from two (germline-reverted IgG and IgM VRC01) or three (mature and germline-reverted IgG and IgM NIH45-46) independent experiments. *, actin-normalized log-transformed signal intensities that are statistically different from the unstimulated controls using a repeated measures ANOVA methodology, followed by Tukey’s multiple comparisons test. (i–k) B cells expressing the mature NIH45-46 (i), germline-reverted NIH45-46 (j), or germline-reverted VRC01 (k) IgG BCR were loaded with Fluo-4 direct calcium indicator and stimulated with the indicated Env proteins. Black arrow indicates time of Env addition. (l) Model illustrating how NIH45-46 (transparent) clashes with glycans at gp120 residues 276 and 460 if glycans are conformationally relaxed in the absence of NIH45-46. (m) Model of NIH45-46 bound to gp120 with glycans removed from 276, 460, and 463 to eliminate potential antibody-glycan clash. Red, gp120; yellow, NIH45-46 contact residues on gp120; blue, NIH45-46 heavy chain; purple, NIH45-46 light chain; orange, glycan at 276; green, glycan at 460; cyan, glycan at 463; white, glycans at other positions. We also examined the interactions between these Envs and glVRC01, a clonal variant of the NIH45-46 antibody (Scheid et al., 2011). Even stronger binding affinities were observed with the glVRC01 antibody (Fig. 2, f and l; and Table S3). In all cases, the increase in the on-rates, rather than decrease in the off-rates, observed when the NLGS are removed from 426c, are suggestive of alleviation of steric constraints rather than changes in epitope conformation. Interestingly, the glNIH45-46 and glVRC01 did not bind the mutant lacking the 460 and 463 NLGS (Fig. 2, h and i). The mutant Envs lacking the 276 NLGS or all three NLGS bound the glNIH45-46 BCR (Fig. 3 a) and activated the corresponding B cells (increase in tyrosine phosphorylation [Fig. 3, c and g] and Ca2+ flux [Fig. 3 j]). In agreement with our binding data for the glVRC01 (Table S3), which indicated stronger Env-binding affinities for glVRC01 than glNIH45-46, the ensuing B cell activation with these two Envs was stronger (Fig. 3, d, g, and k) than that observed with B cells expressing the glNIH45-46 BCR (Fig. 3, c, g, and j). Because the intensity of BCR signaling through the IgG form is stronger than through the IgM form (Wakabayashi et al., 2002; Horikawa et al., 2007; Waisman et al., 2007), we performed our studies with the IgG form, to ensure that any weak signaling induced by our Env proteins would be detected. However, when germline BCRs are expressed on naive B cells, they are of the IgM or IgD classes. We confirmed that the signaling pattern through the IgM glNIH45-46 and glVRC01 BCRs mirrors that observed with the corresponding IgG BCRs, but, as expected, the intensity of the signal is lower (Fig. 3, e, f, and h). The aforementioned results suggest that the NLGS at position 276 plays a critical role in the binding efficiencies of mature and germline-reverted NIH45-46 and VRC01 with Env. Structural modeling suggests that this NLGS hinders antibody binding by interfering with the light antibody chain (Fig. 3 l). Our model indicates that the NLGS at position 463 is not close enough to the antibody epitope to be a major player in this interaction. The model and the improved affinities of germline-reverted and mature NIH45-46 and VRC01 for the triple mutant over the single N276D mutant are consistent with the hypothesis that the glycan at 460 also limits the accessibility of the antibody; the fact that removal of 460 and 463 alone does not significantly improve germline-reverted affinity emphasizes that germline-reverted binding is dependent on removal of the glycan at 276. The CDRL1 of mNIH45-46 and mVRC01 have a two-residue deletion relative to the germline-reverted antibodies (Fig. S1; Zhou et al., 2010; Diskin et al., 2011; Scheid et al., 2011) and this difference may result in differential interactions with the NLGS at positions 276 and 460. Future studies will reveal whether or not similar mutations on different Envs also lead to glNIH45-46 or glVRC01 binding. In accordance with the aforementioned antibody Env-binding results, mNIH45-46 neutralized 426c viruses expressing the deglycosylated Env variants more efficiently than the parental virus (Fig. 4 a), whereas glNIH45-46 weakly neutralized viruses expressing either the single (276 NLGS) or triple NLGS mutant Envs, but not viruses expressing WT or Env lacking the 460 and 463 NLGS (Fig. 4 b). The NLGS at position 276 is highly conserved among circulating HIV-1 strains and only 5.0% of the Env sequences lack this glycosylation site. Interestingly, a higher proportion of clade C than clade B Envs lack this glycosylation site (7.4% of C and 2.0% of B). Even less frequent is the simultaneous absence of all three NLGS (3.1%). Potentially, viral clones with rare glycosylation patterns on Env emerge during infection and stimulate the germline-reverted BCRs of VRC01 class antibodies, such as NIH45-46 and VRC01, thus starting the process of antibody affinity maturation that eventually leads to the development of broadly neutralizing antibody responses (Gray et al., 2011a; Mikell et al., 2011). The rarity of circulating Envs lacking these NLGS may in part explain why VRC01 class antibody responses are elicited only by a minority of those infected with HIV (Lynch et al., 2012). We note that the above NLGS mutations on the backbone of the 426c Env do not diminish the ability of that Env to mediate virus–cell entry (Fig. 4 d). We do not yet know whether all the naturally occurring Envs that lack those three NLGS bind glNIH45-46 and glVRC01. We also do not know whether the modifications we discuss here will always lead to glNIH45-46 or glVRC01 binding when introduced on the backbone of all the Envs we tested (Table S2). Most likely, they will not always result in antibody binding because additional structural elements of Env will vary in a backbone-specific manner and may limit antibody binding even in the absence of the three NLGS discussed here. Structural information of Envs that do and do not bind glNIH45-46 and glVRC01 will be very useful to better understand why some Env are recognized by these antibodies and why some are not. Such structural studies will greatly assist Env-immunogen design efforts that are geared toward stimulating B cells expressing VRC01 class BCRs. Figure 4. Neutralization of 426c viral variants by NIH45-46. (a and b)Entry of pseudovirions expressing either the wild-type 426c Env or derivatives lacking NLGS at positions 276, 460, and 463 into TZM-bl cells was measured in the presence of mature (a) or germline-reverted (b) NIH45-46 IgG. Pseudovirions expressing the unrelated murine leukemia virus (MLV) envelope are included as controls. Entry of the indicated pseudovirions into TZM-bl cells in the presence of mAb PG9 was used as control for nonspecific neutralization that may sometimes occur at high mAb concentrations (c). Each data point represents the mean ± SEM of relative luciferase units of two independent experiments, each done in duplicate. (d) Single-round entry competent pseudovirions expressing the indicated 426c Env were added to TZMbl cells in a 96-well microtiter plate at 0.1 ng/ well p24 (n = 6; open bars), and 1 ng/ well p24 (n = 6; filled bars). Bars represent the mean ± SEM of relative luciferase units expressed relative to that obtained with the wild-type virus (n = 6). Despite the potent binding of other VRC01 class members studied here (12A21, 3BNC60, and 3BNC117; Fig. 5 a) to the 426c Env lacking all three NLGS, their germline-reverted versions did not recognize that Env (Fig. 5 b). The heavy chains of mAbs NIH45-46, VRC01, 12A21, 3BNC60, and 3BNC117 are derived from the same VH1-2*02 allele (Zhou et al., 2010; Scheid et al., 2011). However, the light chains of mAbs NIH45-46 and VRC01 are derived from Vk3-11, whereas those of 12A21, 3BNC60, and 3BNC117 are derived from Vk1D-33. Structural differences between the light chains among these antibodies (Fig. S1) could be responsible for their different Env-recognition properties. Indeed, when the germline-reverted light chains of 12A21 and 3BNC60 were replaced by that of NIH45-46/VRC01, the chimeric germline-reverted antibodies recognized the 426c NLGS mutant Envs (Fig. 5, c and d). Thus, the mutations in loop D and the V5 region identified here may be the minimal set of mutations allowing the binding of germline VRC01 class antibodies to the CD4-BS. It will be important to identify additional Env mutations that alleviate clashes between the germline-reverted light chains of antibodies such as 12A21 or 3BNC60 and elements of Env. Figure 5. Binding of mature and germline-reverted anti–CD4-BS bnmAbs to the 426 Env lacking NLGS at positions 276, 460, and 463. Binding of the mature (a) and germline-reverted (b) antibodies to 426c Env lacking all three NLGS at a concentration of 1 µM was measured by BLI. Binding of the germline-reverted NIH45-46 light chain with the germline-reverted 3BNC60/3BNC117 heavy chain (c) and the germline-reverted NIH45-46 light chain with the germline-reverted 12A21 heavy chain (d) to the indicated trimeric Env was measured by BLI. All Env were tested at concentration of 1 µM. Our experiments indicate that a potential reason for the lack of elicitation of VRC01 antibodies, by Env immunogens is the inability of these immunogens to stimulate B cells expressing the germline BCRs of such antibodies and initiate the maturation process that leads to the production of bNAbs against one of the most conserved HIV Env regions. We show that glycosylation of specific Env regions hinders recognition by the germline-reverted BCRs of two of the most potent VRC01 class antibodies. As such, our findings provide a pathway to overcome a crucial, early, roadblock in the elicitation of similar antibodies by immunization. We note that additional blocks may prevent the elicitation of such antibodies during immunization and that potentially prime boost immunization strategies need to be devised to guide BCR evolution toward specific B cell maturation pathways that are necessary for the production of VRC01 class antibodies (Kwong and Mascola, 2012). MATERIALS AND METHODS Antibodies and BCR construction. The anti–CD4-BS mAbs discussed here were isolated from different HIV-infected subjects by VH+VL gene amplification from single B cells (Wu et al., 2010; Scheid et al., 2011). Soluble IgGs were expressed in 293F cells and purified by Protein A affinity chromatography. Mature and germline-reverted IgG forms of VRC01 were provided by J. Mascola and X. Wu (National Institutes of Health, Bethesda, MD). To generate plasmids expressing the membrane-anchored forms of the anti-HIV antibodies, the following strategy was used. The variable heavy and light antibody regions (separated by a furin cleavage site and an F2A peptide sequence) were inserted into pTT5 vector. The constant IgG and IgM splice variant cDNAs (containing the two exons that encode the transmembrane and cytoplasmic domains of human IgG and IgM) were synthesized by GenScript. That sequence was introduced 3′ to the variable heavy region in the aforementioned pTT5 vector. These vectors express the membrane-anchored version of either the mature or germline-reverted versions of the antibodies studied here. Cells. The DG-75 human Burkitt’s lymphoma (#CRL-2625; ATCC) cell line was maintained in RPMI-1640 supplemented with 10% FBS. DG-75 cells express human IgM do not express Fc receptors. Neutralization assays. Neutralization assays were performed as previously described (Saunders et al., 2005). TZM-bl cells were maintained in DMEM (Cellgro) supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. For neutralization assays, the cells were plated at a density of 5 × 104 cells per well 24 h before addition of virus and antibody. Antibodies were serially diluted in microtiter wells at the indicated starting concentration in a total volume of 30 µl per well. 30 µl of virus (previously determined to result in 2 × 105 luciferase units) was added to each well containing the titrated antibodies. Antibody-virus mixtures in duplicate wells were incubated at 37°C for 1.5 h. During the last 30 min of the incubation, cells were treated at 37°C with 1 µg/ml of polybrene. The polybrene was then aspirated from the cells and replaced with 50 µl of antibody-pseudovirus mixture. Plates were incubated at 37°C for 72 h, media was aspirated, and cells were lysed to measure luciferase activity using Steady-Glo Luciferase Reagent (Promega) ELISA. Env ELISA assays were performed with established methodologies, either by directly adsorbing recombinant Env (SIVmac239, JRCSF, BAL, Consensus C, DU151, Consensus A1, MJ613, and A244; Immune Technologies) to ELISA plates or by capturing p120 released from pseudoviruses after lysis by 1% Triton X-100 on D7324-coated (anti–C-terminal gp120 sheep antibody) ELISA wells. In the first case, the binding cut off for ELISA was defined as the mean + 3 SD of A450 recorded with the SIVmac gp120 at the highest antibody concentration, whereas in the second case the binding cut off was the mean + 3 SD of the absorbance obtained for each antibody against the capture antibody alone (no Env). Biolayer interferometry analysis of mAb-Env binding. These assays were performed on the Octet KQe instrument (ForteBio, Inc.). IgGs (20 µg/ml in PBS) were immobilized onto anti–human IgG FC capture (AHC) biosensors (ForteBio) for 5 min. The baseline interference was then read for 60 s, in kinetics buffer (KB: 1× PBS, 0.01% BSA, 0.02%, Tween 20, and 0.005% NaN3),followed by subsequent immersion of the sensors into wells containing recombinant trimeric Env gp140 diluted in KB for 300s (association phase). Sensors were then immersed in KB for the indicated times (dissociation phase). All kinetic interactions were measured with new sensors at 30°C and shaking at 1,000 RPM in 96-well plates. Curve fitting was done using a 1:1 binding model using the Data analysis software (ForteBio) and using the concentration of gp140 protomers in solution. Mean k on and k off and apparent K d values were determined from all binding curves that matched the theoretical fit with an R2 value of ≥0.95. HIV envelopes. The following Envs were examined: clade A: Q168a2, Q259d2, Q461e2, Q769h5; clade B: SF162, SF162ΔV1, SF162ΔV2, SF162ΔV3; clade C: 823c, 756c, 706c, 459c, 140c, 327c, 405c and 426c. Proteins were either purchased or expressed and purified in-house as previously described (Sellhorn et al., 2009). These clade C Envs (available from under GenBank accession nos. KC769511–KC769518) were derived from acutely infected participants in the HVTN503 clinical trial (Gray et al., 2011b). The JRCSF, SF162, ADA, JRFL, YU2, and HXB2 Envs were derived from clade B viruses isolated during chronic infection. The clade C Envs Du151, Du422, CAP45, and ZM249 and the clade B Envs QH0692, AC10.0, PVO, TRO, RHPA, TRJO, WITO, REJO, and THRO were derived from viruses isolated during the first 6 mo of infection. The clade A Envs Q168a2, Q259d2, Q461e2, and Q769h5 were derived from viruses isolated between 28 and 75 d after infection. The A244 Env was derived from a clade A/E virus. Expression and activation of exogenous BCRs on B cells. 2 × 106 DG-75 B cells in 100 µl of cell line Nucleofector solution V (Lonza) were electroporated with 5 µg of BCR-expressing plasmids (Amaxa Nucleofector II; Lonza; program O-006). Cell surface expression of exogenous human BCRs was determined by allophycocyanin (APC)-conjugated mouse monoclonal anti–human IgG (1/10; BD). 24 h after transfection, cells were loaded with Fluo-4 Direct calcium indicator (Invitrogen), in RPMI-1640 medium containing 10% FBS at 37°C for 45 min. Cells were pelleted and stained with APC-conjugated mouse monoclonal anti–human IgG (1/10 dilution in 100 µl of RPMI-1640 with 10% FBS and Fluo-4 Direct) for 15 min. The cells were washed with 5 ml in RPMI-1640 containing 10% FBS, pelleted, and resuspended at ∼106 cells/ml in RPMI-1640 and subjected to Ca2+ flux analysis at a medium flow rate on an LSR II cytometer (BD). For all the experiments, we gated on B cells expressing comparable numbers of BCRs. Minimum levels of background fluorescence (MinFL) were determined by averaging the background Fluo-4 absorbance in cells for 30 s. After that, activation of exogenous BCRs by recombinant Env (30–400 µg/ml) was determined by monitoring changes in Fluo-4 fluorescence associated with cells expressing the exogenous BCRs (APC positive cells) for 210 or 270 s. Ionomycin was added to a final concentration of 6.5 nM for 60s and maximum Fluo-4 fluorescence (MaxFL) was established by averaging changes in Fluo-4 fluorescence recorded during the last 10 s. The percentage of maximum Fluo-4 fluorescence at each time point t was determined using the formula: (Fluorescence at t-MinFL) / (MaxFL-MinFL) × 100. This analysis was performed on both the BCR-positive (anti–IgG-APC–positive) and BCR-negative (anti–IgG-APC–negative) cells simultaneously. The background Fluo-4 fluorescence signal from the BCR-negative cells was subtracted from that of the BCR positive population at each time point. Tyrosine phosphorylation activity after a 3-min incubation at room temperature with Env (40 or 200 µg/ml) was determined in DG75 cells expressing NIH45-46 and VRC01 BCRs. Cells were pelleted and resuspended in lysis buffer (1% NP-40, 10% glycerol, 2 mM EDTA, 137 mM NaCl, and 20 mM Tris HCl, pH 8.0) supplemented with phosphatase inhibitors (Halt Phosphatase Inhibitor Cocktail; Thermo Fisher Scientific) for 10 min on ice. Lysates were cleared by centrifugation and subjected to SDS-PAGE and Western blot analysis using 4G10 pY mAb (Millipore) and Anti–β-actin rabbit mAb (13E5; Cell Signaling Technology). Bands were visualized with IR-dye conjugated secondary antibodies and imaged and quantified by Odyssey Infrared Imaging System (LI-COR Biosciences). Env sequence analysis. The 2010 Filtered Web Alignment (excluding 4 SIV sequences; Los Alamos HIV Database, http://www.hiv.lanl.gov/) was used to determine the frequency of NLGS in Env. Recombinant Env binding to exogenous BCRs expressed on B cells. BCR expressing and mock transfected DG-75 cells were stained with APC-conjugated mouse anti–human IgG, as described above. After washing in PBS containing 10% FBS (FACS wash buffer) cells were resuspended (106 cells/ml) in FACS wash buffer and 1.0 × 105 cells were incubated with 30 µg/ml of gp140 Env for 20 min on ice. The cells were washed and incubated with R-phycoerythrin (PE)–labeled IgG purified from pooled sera of 50 HIV-1+ subjects. After centrifugation and washing (2× in ice cold FACS wash buffer), the cells were resuspended in 200 µl of 1% paraformaldehyde and analyzed on an LSR II cytometer (BD). Envelope surface staining was expressed as the MFI of PE staining in the presence of Env + HIV Ig PE minus the MFI obtained with HIV-IgG-PE alone on BCR expressing (APC-positive) cells. Env mutagenesis. Point mutations in the gp140 and gp160 envelope sequences were introduced by site-directed mutagenesis (Stratagene Quick Change II system from Agilent Technologies, Santa Clara, California), using the following primers (sense strand provided) 426c-N460D/N463D (5′-GATGGGGGAAACACTACCGATAACACCGAGATTTCC-3′) and 426c-N276D (5′-GAGGAAGAGATTGTGATCAGATCAAAAGACCTGAGCGATAAT-3′). Modeling the NIH45-46-Env interaction. A homology model for core gp120 of clade C strain 426c was built by RosettaRemodel (Huang et al., 2011), using the NIH45-46-gp120 crystal structure (PDB accession no. 3u7y) as a structural template. Low-energy backbone conformations were generated for the V5 loop in the presence of bound NIH45-46. Representative Man8GlcNac2 glycans were then aligned onto each gp120 Asn residue within a glycosylation sequon, and low-energy glycan conformations were modeled using GlycanRelax (Schief et al., 2009). Statistical analysis. Significant increase of tyrosine phosphorylation was assessed by repeated measures ANOVA followed by Tukey’s multiple comparisons test comparing log-transformed, actin-normalized Western blot signal intensities of antigen-stimulated cells with that of unstimulated cells. A p-value <0.05 was considered significant. Online supplemental material. Fig. S1 shows amino acid alignment of the germline-reverted antibody heavy and light chains. Table S1 shows characteristics of VRC01 class antibodies. Table S2 shows IgG binding to recombinant Env proteins. Table S3 shows the binding kinetics of mature and germline NIH 45-46 IgG to trimeric 426c Env gp140 variants. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20122824/DC1. Supplementary Material Supplemental Material