- Record: found
- Abstract: found
- Article: found
From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo
Read this article at
Abstract
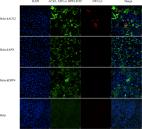
The human-animal-environment interface of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an important aspect of the coronavirus disease 2019 (COVID-19) pandemic that requires robust One Health-based investigations. Despite this, few reports describe natural infections in animals or directly link them to human infections using genomic data. In the present study, we describe the first cases of natural SARS-CoV-2 infection in tigers and lions in the United States and provide epidemiological and genetic evidence for human-to-animal transmission of the virus. Our data show that tigers and lions were infected with different genotypes of SARS-CoV-2, indicating two independent transmission events to the animals. Importantly, infected animals shed infectious virus in respiratory secretions and feces. A better understanding of the susceptibility of animal species to SARS-CoV-2 may help to elucidate transmission mechanisms and identify potential reservoirs and sources of infection that are important in both animal and human health.
ABSTRACT
Despite numerous barriers to transmission, zoonoses are the major cause of emerging infectious diseases in humans. Among these, severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and ebolaviruses have killed thousands; the human immunodeficiency virus (HIV) has killed millions. Zoonoses and human-to-animal cross-species transmission are driven by human actions and have important management, conservation, and public health implications. The current SARS-CoV-2 pandemic, which presumably originated from an animal reservoir, has killed more than half a million people around the world and cases continue to rise. In March 2020, New York City was a global epicenter for SARS-CoV-2 infections. During this time, four tigers and three lions at the Bronx Zoo, NY, developed mild, abnormal respiratory signs. We detected SARS-CoV-2 RNA in respiratory secretions and/or feces from all seven animals, live virus in three, and colocalized viral RNA with cellular damage in one. We produced nine whole SARS-CoV-2 genomes from the animals and keepers and identified different SARS-CoV-2 genotypes in the tigers and lions. Epidemiologic and genomic data indicated human-to-tiger transmission. These were the first confirmed cases of natural SARS-CoV-2 animal infections in the United States and the first in nondomestic species in the world. We highlight disease transmission at a nontraditional interface and provide information that contributes to understanding SARS-CoV-2 transmission across species.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
A Novel Coronavirus from Patients with Pneumonia in China, 2019

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability
- Record: found
- Abstract: found
- Article: found