- Record: found
- Abstract: found
- Article: not found
Metagenomic survey of antimicrobial resistance (AMR) in Maryland surface waters differentiated by high and low human impact
brief-report

Brandon Kocurek
1 ,
Shawn Behling
2 ,
Gordon Martin
1 ,
Padmini Ramachandran
3 ,
Elizabeth Reed
3 ,
Christopher Grim
3 ,
Mark Mammel
3 ,
Jie Zheng
3 ,
Alison Franklin
4 ,
Jay Garland
4 ,
Daniel A. Tadesse
1 ,
Manan Sharma
5 ,
Gregory H. Tyson
1 ,
Claudine Kabera
1 ,
Heather Tate
1 ,
Patrick F. McDermott
1 ,
Errol Strain
1 ,
Andrea Ottesen
1
,
January 2024
30 November 2023
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
ABSTRACT
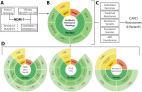
Here, we examine surface waters as a modality to better understand baseline antimicrobial resistance (AMR) across the environment to supplement existing AMR monitoring in pathogens associated with humans, foods, and animals. Data from metagenomic and quasimetagenomic (shotgun sequenced enrichments) are used to describe AMR in Maryland surface waters from high and low human impact classifications.
Related collections
Most cited references10
- Record: found
- Abstract: found
- Article: found
CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database
Brian P. Alcock, Amogelang Raphenya, Tammy Lau … (2019)

- Record: found
- Abstract: found
- Article: found
AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence

- Record: found
- Abstract: found
- Article: found
MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data
Enrique Doster, Steven Lakin, Christopher J. Dean … (2019)
