- Record: found
- Abstract: found
- Article: found
The IKAROS Interaction with a Complex Including Chromatin Remodeling and Transcription Elongation Activities Is Required for Hematopoiesis
Read this article at
Abstract
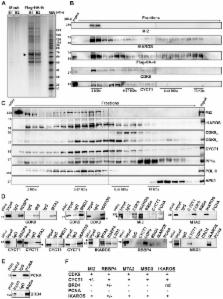
IKAROS is a critical regulator of hematopoietic cell fate and its dynamic expression pattern is required for proper hematopoiesis. In collaboration with the Nucleosome Remodeling and Deacetylase (NuRD) complex, it promotes gene repression and activation. It remains to be clarified how IKAROS can support transcription activation while being associated with the HDAC-containing complex NuRD. IKAROS also binds to the Positive-Transcription Elongation Factor b (P-TEFb) at gene promoters. Here, we demonstrate that NuRD and P-TEFb are assembled in a complex that can be recruited to specific genes by IKAROS. The expression level of IKAROS influences the recruitment of the NuRD-P-TEFb complex to gene regulatory regions and facilitates transcription elongation by transferring the Protein Phosphatase 1α (PP1α), an IKAROS-binding protein and P-TEFb activator, to CDK9. We show that an IKAROS mutant that is unable to bind PP1α cannot sustain gene expression and impedes normal differentiation of Ik NULL hematopoietic progenitors. Finally, the knock-down of the NuRD subunit Mi2 reveals that the occupancy of the NuRD complex at transcribed regions of genes favors the relief of POL II promoter-proximal pausing and thereby, promotes transcription elongation.
Author Summary
Perturbation of the expression level of IKAROS, a transcription factor critical during hematopoiesis, is associated with malignant transformation in mice and humans. The importance of IKAROS expression levels for the control of target-gene regulation was addressed in hematopoietic progenitor cells. The collaboration between IKAROS and the Nucleosome Remodeling and Deacetylase (NuRD) complex can promote gene activation or repression. IKAROS can also interact with the Positive-Transcription Elongation Factor b (P-TEFb) and the Protein Phosphatase 1 (PP1), an important P-TEFb regulator. Immunoaffinity purification of IKAROS interacting proteins and Fast Protein Liquid Chromatography analysis revealed a dynamic interaction between IKAROS, PP1 and the newly defined NuRD-P-TEFb complex. This complex can be targeted to specific genes in cells expressing high levels of IKAROS to promote productive transcription elongation. Based on our results we suggest that, in addition to P-TEFb, the NuRD complex and PP1 are required to facilitate transcription elongation of IKAROS-target genes and normal differentiation of hematopoietic progenitor cells.
Related collections
Most cited references65
- Record: found
- Abstract: found
- Article: not found
Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells.
- Record: found
- Abstract: found
- Article: not found
The super elongation complex (SEC) family in transcriptional control.
- Record: found
- Abstract: found
- Article: not found