- Record: found
- Abstract: found
- Article: found
Symbioses are restructured by repeated mass coral bleaching

Read this article at
Abstract
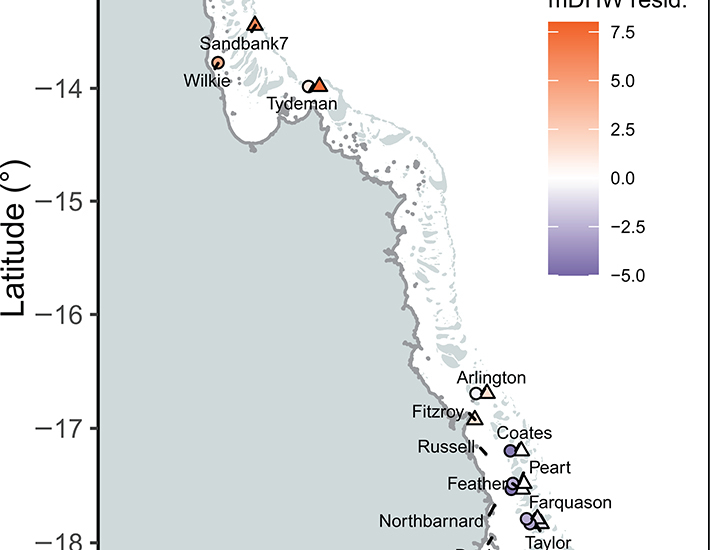
Survival of symbiotic reef-building corals under global warming requires rapid acclimation or adaptation. The impact of accumulated heat stress was compared across 1643 symbiont communities before and after the 2016 mass bleaching in three coral species and free-living in the environment across ~900 kilometers of the Great Barrier Reef. Resilient reefs (less aerial bleaching than predicted from high satellite sea temperatures) showed low variation in symbioses. Before 2016, heat-tolerant environmental symbionts were common in ~98% of samples and moderately abundant (9 to 40% in samples). In corals, heat-tolerant symbionts were at low abundances (0 to 7.3%) but only in a minority (13 to 27%) of colonies. Following bleaching, environmental diversity doubled (including heat-tolerant symbionts) and increased in one coral species. Communities were dynamic ( Acropora millepora) and conserved ( Acropora hyacinthus and Acropora tenuis), including symbiont community turnover and redistribution. Symbiotic restructuring after bleaching occurs but is a taxon-specific ecological opportunity.
Abstract
Abstract
Climate change-driven mass coral bleaching restructures algal symbiont communities in some corals and in the environment.
Related collections
Most cited references62
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data

- Record: found
- Abstract: found
- Article: found
phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data
- Record: found
- Abstract: not found
- Article: not found