- Record: found
- Abstract: found
- Article: found
The challenges of COVID‐19 Delta variant: Prevention and vaccine development

Abstract
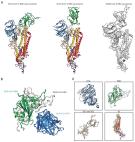
Several SARS‐CoV‐2 variants have emerged since the pandemic, bringing about a renewed threat to the public. Delta variant (B.1.617.2) was first detected in October 2020 in India and was characterized as variants of concern (VOC) by WHO on May 11, 2021. Delta variant rapidly outcompeted other variants to become the dominant circulating lineages due to its clear competitive advantage. There is emerging evidence of enhanced transmissibility and reduced vaccine effectiveness (VE) against Delta variant. Therefore, it is crucial to understand the features and phenotypic effects of this variant. Herein, we comprehensively described the evaluation and features of Delta variant, summarized the effects of mutations in spike on the infectivity, transmission ability, immune evasion, and provided a perspective on efficient approaches for preventing and overcoming COVID‐19.
Abstract
SARS‐CoV Delta (B.1.617.2) variant has been classified as variants of concern (VOC) by World Health Organization (WHO). The reproductive number (R0) of SARS‐CoV‐2 wild type and Delta variant is 2.3‐5.7 and 5‐8, respectively. Patients infected by Delta variant exhibit shorter mean generation time and mean serial interval, higher virus load and hospitalization rate compared to those infected by wild‐type SARS‐CoV‐2.
Related collections
Most cited references82
- Record: found
- Abstract: found
- Article: not found
Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia
- Record: found
- Abstract: found
- Article: not found
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding
- Record: found
- Abstract: found
- Article: found
