- Record: found
- Abstract: found
- Article: found
The soft explosive model of placental mammal evolution
Read this article at
Abstract
Background
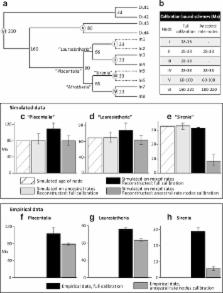
Recent molecular dating estimates for placental mammals echo fossil inferences for an explosive interordinal diversification, but typically place this event some 10–20 million years earlier than the Paleocene fossils, among apparently more “primitive” mammal faunas.
Results
However, current models of molecular evolution do not adequately account for parallel rate changes, and result in dramatic divergence underestimates for large, long-lived mammals such as whales and hominids. Calibrating among these taxa shifts the rate model errors deeper in the tree, inflating interordinal divergence estimates. We employ simulations based on empirical rate variation, which show that this “error-shift inflation” can explain previous molecular dating overestimates relative to fossil inferences. Molecular dating accuracy is substantially improved in the simulations by focusing on calibrations for taxa that retain plesiomorphic life-history characteristics. Applying this strategy to the empirical data favours the soft explosive model of placental evolution, in line with traditional palaeontological interpretations – a few Cretaceous placental lineages give rise to a rapid interordinal diversification following the 66 Ma Cretaceous-Paleogene boundary mass extinction.
Conclusions
Our soft explosive model for the diversification of placental mammals brings into agreement previously incongruous molecular, fossil, and ancestral life history estimates, and closely aligns with a growing consensus for a similar model for bird evolution. We show that recent criticism of the soft explosive model relies on ignoring both experimental controls and statistical confidence, as well as misrepresentation, and inconsistent interpretations of morphological phylogeny. More generally, we suggest that the evolutionary properties of adaptive radiations may leave current molecular dating methods susceptible to overestimating the timing of major diversification events.
Related collections
Most cited references58
- Record: found
- Abstract: found
- Article: not found
The delayed rise of present-day mammals.
- Record: found
- Abstract: found
- Article: not found
The placental mammal ancestor and the post-K-Pg radiation of placentals.
- Record: found
- Abstract: found
- Article: not found