- Record: found
- Abstract: found
- Article: found
Automated assembly of a reference taxonomy for phylogenetic data synthesis
research-article
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Introduction
Any large biodiversity data project requires one or more taxonomies for discovery
and data integration purposes, as in "find occurrence records for primates" or "find
the taxon record associated with this sequence" (Page 2008). Examples of such projects
are GBIF (Edwards 2004), which focuses on occurrence records, and NCBI (Federhen 2011),
which focuses on genetic sequence records. Each of these projects has a dedicated
taxonomy effort that is responsive to the project's particular needs. We present the
design and application of the Open Tree Taxonomy, which serves the Open Tree of Life
project, an aggregation of phylogenetic trees with tools for operating on them. (Hinchliff
et al. 2015
McTavish et al. 2015, Redelings and Holder 2017, Open Tree of Life project 2017).
In order to meet Open Tree's project requirements, the taxonomy is an automated assembly
of ten different source taxonomies. The assembly process is repeatable so that we
can easily incorporate updates to source taxonomies. Repeatability also allows us
to easily test potential improvements to the assembly method. Information about taxa
is typically expressed in databases and files in terms of taxon names or 'name-strings'
(Patterson 2014). To combine taxonomies it is therefore necessary to be able to determine
name equivalence: whether or not an occurrence of a name-string in one data source
refers to the same taxon as a given name-string occurrence in another. Solving this
equivalence problem requires that we distinguishing occurrences that only coincidentally
have the same name-string (homonym sense detection), and unify occurrences only when
evidence justifies it. We have developed a set of heuristics that scalably address
this equivalence problem.
The Open Tree of Life project
The Open Tree of Life project consists of a set of tools for:
synthesizing summary phylogenetic trees ('synthetic trees') from a corpus of phylogenetic
tree inputs (input trees)
matching groupings in synthetic trees with higher taxa (such as
Mammalia
)
supplementing synthetic trees with taxa obtained only from taxonomy.
The outcome is one or more synthetic trees combining phylogenetic and taxonomic knowledge.
Fig. 1 illustrates an overview of the process of combining phylogenetic trees and
taxonomy, while the details are described in a separate publication (Redelings and
Holder 2017).
Although Open Tree is primarily a phylogenetic tree aggregation effort, it requires
a reference taxonomy that can support each of these functions.
For synthetic tree synthesis (1), we use the taxonomy for converting OTUs (operational
taxonomic units, or 'tips') on input trees to a canonical form. Synthetic tree construction
requires that an input tree OTU be matched with an OTU from another input tree when,
and only when, it is reasonable to do so. This is a nontrivial task because a taxon
can have very different OTU labels in different input trees due to synonymies, abbreviations,
misspellings, notational differences, and so on. In addition, a given label can name
different taxa in different trees (homonymy). The approach we take is to map OTUs
to the reference taxonomy, so that OTUs in different input trees are compared by comparing
the taxa to which they map.
For higher taxon associations (2), we compare the groupings in the synthetic tree
to those in the taxonomy.
For supplementation (3), only a relatively small number of described taxa are represented
in input trees (currently about 200,000 in the phylogenetic corpus out of two million
or more known taxa), so the taxonomy provides those that are not. The large complement
of taxonomy-only taxa can be 'grafted' onto a synthetic tree in phylogenetically plausible
locations based on how they relate taxonomically to taxa that are known from input
trees.
Reference taxonomy requirements
This overall program dictates what we should be looking for in a reference taxonomy.
In addition to the technical requirements derived from the above, we have two additional
requirements coming from a desire to situate Open Tree as ongoing infrastructure for
the evolutionary biology community, rather than as a one-off study. Following are
all five requirements:
OTU coverage: The reference taxonomy should have a taxon at the level of species or
higher for every OTU that has the potential to occur in more than one study, over
the intended scope of all cellular organisms.
Phylogenetically informed classification: Higher taxa should be provided with as much
resolution and phylogenetic fidelity as is reasonable. Ranks and nomenclatural structure
should not be required (since many well-established groups do not have proper Linnaean
names or ranks) and groups at odds with phylogenetic understanding (such as
Protozoa
) should be avoided.
Taxonomic coverage: The taxonomy should cover as many as possible of the species that
are described in the literature, so that we can supplement synthetic trees as described
in step 3 above.
Ongoing update: New taxa of importance to phylogenetic studies are constantly being
added to the literature. The taxonomy needs to be updated with new information on
an ongoing basis.
Open data: The taxonomy must be available to anyone for unrestricted use. Users should
not have to ask permission to copy and use the taxonomy, nor should they be bound
by terms of use that interfere with further reuse.
An additional goal is that the process should be reproducible and transparent. Given
the source taxonomies, we should be able to regenerate the taxonomy, and taxon records
should provide information about the taxonomic sources from which it is derived.
No single available taxonomic source meets all requirements. The NCBI taxonomy has
good coverage of OTUs, provides a rich source of phyogenetically informed higher taxa,
and is open, but its taxonomic coverage is limited to taxa that have sequence data
in GenBank (only about 360,000 NCBI species having standard binomial names at the
time of this writing). Traditional all-life taxonomies such as Catalogue of Life,
IRMNG (Rees 2008), and GBIF meet the taxonomic coverage requirement, but miss many
OTUs from our input trees, and their higher-level taxonomies are often not as phylogenetically
informed or resolved as the NCBI taxonomy. At the very least, Open Tree needs to combine
an NCBI-like sequence-aware taxonomy with a traditional broad taxonomy that is also
open.
These requirements cannot be met in an absolute sense; each is a 'best effort' requirement
subject to availability of project resources.
Note that the Open Tree Taxonomy is not supposed to be a reference for nomenclature;
it links to other sources for nomenclatural and other information. Nor is it a place
to deposit curated taxonomic information. The taxonomy has not been vetted in detail,
as doing so would be beyond the capacity and focus of the Open Tree project. It is
known to contain many taxon duplications and technical artifacts. Tolerating these
shortcomings is a necessary tradeoff in attempting to meet the above requirements.
Related work
There are probably about a dozen public all-life taxonomy compilations. The methods
of assembly and curation are documented for only a few of these. We assume that most
are evolving databases that are extended and maintained by a combination of single-record
operations and some amount of ad hoc scripting to import material in bulk. The NCBI
taxonomy, described in Federhen 2011, is of this type. Catalogue of Life, which documents
its method in Species 2000 2017, is different in that it has a divide and conquer
approach: it is assembled through systematic grafting of sub-taxonomies received from
a network of editors. Because its sub-taxonomies are nonoverlapping, CoL is not a
synthesis in the sense used here. The GBIF backbone taxonomy is assembled via automated
synthesis of overlapping sub-taxonomies, and in that respect is similar to OTT. The
GBIF method is as yet unpublished, although some information is available (Döring
2016b, Döring 2016a). Note that OTT builds on GBIF, which in turn builds on CoL.
Had an available all-life taxonomy met all of Open Tree's requirements, the project
would have used it. Unfortunately, for each one, there is at least one Open Tree requirement
that goes beyond what the taxonomy provides.
An important part of any synthesis method is name-string parsing and matching, and
this is the focus of the Global Names Architecture (Pyle 2016). In the phase of work
reported here, name parsing and matching are not a priority issue, since exact matches
together with synonym records from the source taxonomies provide a provisional solution
that has been adequate so far. Going forward, matching will need more attention, and
components of the GNA will probably play a role.
The Taxonomy Tree Tool (TTT, Lin et al. 2016) employs a tree merge method similar
to the one used here. It seems to be aimed at assisting manual analysis of tree differences.
It is not clear from available documentation how one would use it to resolve conflicts
in an automated workflow.
Method
The conventional approach to meeting the requirements stated in the introduction would
be to create a database, copy the first taxonomy into it, then somehow merge the second
taxonomy into that, repeating for further sources if necessary. However, it is not
clear how to meet the ongoing update requirement under this approach. As the source
taxonomies change, we would like for the combined taxonomy to contain only information
derived from the latest versions of the sources, without residual information from
previous versions. Many changes to the sources are corrections, and we do not want
to retain information that has been corrected or superseded by a later version of
a source.
Rather than maintain a database of taxonomic information, we instead developed a process
for assembling a taxonomy from two or more taxonomic sources. With a repeatable process,
we can generate a new combined taxonomy version from new source taxonomy versions
de novo, and do so frequently. There are additional benefits as well, such as the
ability to add new sources relatively easily, and to use the tool for other purposes.
In the following, any definite claims or measurements refer to the Open Tree reference
taxonomy version 3.0.
Terminology
source taxonomy: imported taxonomic source (NCBI taxonomy, etc.)
workspace: data structure for creation of the reference taxonomy
name-string: name of one or more taxa, without author information, considered as a
sequence of characters, without association with any particular description, or nomenclatural
code
node: a data record intended to correspond to a taxon. Records name-strings, authorship,
parent node, optional rank, optional annotations. If a workspace node, it originates
from a single source taxonomy, and records its source (provenance) and its alignments
to others
parent (node): the nearest enclosing node within a given node's taxonomy
tip: a node that is not the parent of any node
primary name-string: one particular name-string of a node. Each node has exactly one
primary name-string
homonym name-string: a name-string that belongs to multiple nodes within the same
taxonomy. This is analogous to the nontechnical meaning of 'homonym' and is not to
be confused with 'homonym' in the nomenclatural sense, which only applies within a
single nomenclatural code. Nomenclatural homonyms and hemihomonyms (Shipunov 2011)
both correspond to homonym name-strings, as do clerical errors where multiple nodes
are created for the same taxon
synonym name-string (of a node): a non-primary name-string
image (of a node n'): the workspace node corresponding to n'
incertae sedis: node A is incertae sedis in node B if A is a child of B but is not
known to be disjoint (as a taxon) from B's non-incertae-sedis children. That is, if
we had more information, it might turn out that A is a member of one of the other
children of B.
Method overview
This section is an overview of the taxonomy assembly method. Several generalities
stated here are simplifications; the actual method (described later) is significantly
more involved.
We start with a sequence of source taxonomies S1, S2, ..., Sn, ordered by priority.
Priority is the means by which conflicts between sources are resolved, and therefore
has a profound effect on the outcome of assembly. If a curator judges S to be more
accurate or otherwise "better" than S', then S will occur earlier in the priority
sequence than S' and its information supersedes that from later sources. Curators
(either project personnel or participants in Open Tree workshops and online forums)
determine priority based on their taxonomic expertise. Source taxonomies are sometimes
split into pieces in order to establish different priorities for different parts.
Priority choice by curator is a fragile and subjective aspect of the method, but we
could not identify any other information available at scale that could be brought
to bear on conflict resolution.
We define an operator for combining taxonomies pairwise, written schematically as
U = S + S', and apply it from left to right:
U0 = empty, U1 = U0 + S1, U2 = U1 + S2, U3 = U2 + S3...
The combination S + S' is formed in two steps:
A mapping or alignment step that identifies all nodes in S' that can be equated with
nodes in S. There will often be nodes in S' that cannot be aligned to S.
A merge step that creates the combination U = S + S', by adding to S the unaligned
taxa from S'. The attachment position of unaligned nodes from step 1 is determined
from nearby aligned nodes, either as a graft or an insertion.
Examples of these two cases are given in Figure 2.
As a simple example, consider a genus represented in both taxonomies, but containing
different species in the two:
S = (b,c,d)a, S' = (c,d,e)a
S and S' each have four nodes. Suppose c, d, and a in S' are aligned to c, d, and
a in S. The only unaligned node is e, which is a sibling of c and d and therefore
grafted as a child of a. After the merge step, we have:
S + S' = (b,c,d,e)a
One might call this merge heuristic 'my sibling's sibling is my sibling' or 'transitivity
of siblinghood'.
This is a very common pattern. Fig. 2 illustrates a real life-example when combining
the genus
Bufo
across NCBI and GBIF. There are about 900,000 similar simple grafting events in the
assembly of OTT.
The other merge method is an insertion, where the unaligned node has descendants that
are in S. This always occurs when S' has greater resolution than S. For example, see
Fig. 2, where WoRMS provides greater resolution than NCBI.
The vast majority of alignment and merge situations are simple, similar to the examples
shown in Fig. 2. However, even a small fraction of special cases can add up to thousands
when the total number of alignments and merges measures in the millions, so we have
worked to develop heuristics that handle the most common special cases. Ambiguities
caused by homonym name-strings create most of the difficulties, with inconsistent
or unclear higher taxon membership creating the rest. The development of the assembly
process described here has been a driven by trial and error - finding cases that fail
and then adding or modifying alignment heuristics and other logic to address the underlying
cause. Because the sources are noisy and inconsistent, any automated assembly process
will make mistakes. To prevent or correct these mistakes, manual ad hoc adjustments
are applied as needed, as a last resort. The goal in method development is to keep
the number of needed adjustments small.
Taxonomic sources
We build the taxonomy from ten sources. Some of these sources are from taxonomy projects,
while others were manually assembled based on recent publications. As described above,
OTT assembly is dependent on the input order of the sources - higher ranked inputs
take priority over lower ranked inputs. Table 1 lists the sources used to construct
OTT. The full provenance details, and a copy of the normalized source, are available
in supplementary data.
Open Tree curation : It is not uncommon to have taxa as OTUs in phylogenetic studies
that do not occur in OTT. This can be due to a delay in curation by the source taxonomy,
a delay in importing a fresh source version into OTT, a morphological study containing
otherwise unknown species, or other causes. To handle this situation, we developed
a user interface that allows curators to create new taxon records along with relevant
documentation (publications, databases, and so on). New taxon records are saved into
a public GitHub repository, and these records are then linked from the OTT taxonomy
files and user interfaces so that provenance is always available.
Separation taxa : This is a small curated tree containing 31 major groups such as
Fungi
,
Metazoa
, and
Lepidoptera
. Its purpose is to assist in alignment, where homonym name-strings are, or might
need to be, present. If a node is found in one of these separation groups, then it
will not match a node in a disjoint separation group, absent other evidence (details
below).
ARB-SILVA taxonomy processing : The terminal taxa in the SILVA taxonomy are algorithmically
generated clusters of RNA sequences derived from GenBank records. Rather than incorporate
these idiosyncratic, fine-grained groupings into OTT, we use sequence record metadata
to place the clusters into larger groups corresponding to NCBI taxa, and include those
larger groups in OTT.
We excluded SILVA's plant, animal, and fungal branches from OTT because these groups
are well covered by other sources and poorly represented in SILVA. For example, SILVA
has only 299 taxa in
Metazoa
, compared with over 500,000 taxa under
Metazoa
in NCBI Taxonomy.
Extinct / extant annotations : Curators requested information about whether taxa were
extinct vs. extant. With the exception of limited data from WoRMS and Index Fungorum,
this information was not explicitly present in our other sources, so we imported IRMNG,
which logs the extinct / extant status of taxa.
As a secondary heuristic, records from GBIF that originate from PaleoDB, and do not
come from any other taxonomic source, are annotated extinct. This is not completely
reliable, as some PaleoDB taxa are extant.
Suppressed records : We suppress the following source taxonomy records:
animals, plants,
fungi
in SILVA
GBIF backbone records that originate from IRMNG (IRMNG is imported separately)
GBIF backbone records that originate from IPNI
GBIF backbone records whose taxonomic status is 'doubtful'
GBIF backbone records for infraspecific taxa (subspecies, variety, form)
IRMNG records whose nomenclatural status is 'nudum', 'invalid', or any of about 25
similar designations
NCBI Taxonomy records that have no potential for unification with OTUs in phylogenetic
studies: those with name-strings containing 'insertion sequences', 'artificial librarries',
'transposons', or any of about 15 similar designations
The IPNI and IRMNG derived GBIF records are suppressed because they include many invalid
names. We pick up most of the valid names from other sources, such as the direct IRMNG
import, so this is not a great loss. Although GBIF's original taxonomic sources indicate
which names are known to be invalid, this information is not provided by the GBIF
backbone. Note that the GBIF backbone might import the same name from more than one
source, but its provenance information only lists one of the sources. We suppress
the record if that one source is IPNI or IRMNG.
Sources not included : The number of sources was of course limited by the amount of
time we had available for import efforts; new sources were only added for specific
reasons related to curators' interests. The choice of sources was also limited by
the open data goal. Certain obvious choices, such as Catalog of Life, had to be passed
over because there was no access, or access was controlled by legal terms of use.
Import and Normalization
Each source taxonomy has its own import procedure, usually a file download from the
provider's web site followed by application of a script that converts the source to
a common internal form for import (a set of nodes, see terminology section). Given
the converted source files, the taxonomy can be read by the OTT assembly procedure.
After each source taxonomy is loaded, the following normalizations are performed:
Diacritics removal - accents, umlauts, and other diacritic marks are removed in order
to improve name matching, as well as to follow the nomenclatural codes, which prohibit
them. The original name-string is kept.
Child taxa of "containers" in the source taxonomy are made to be children of the container's
parent. "Containers" are groupings in the source that don't represent taxa, for example
nodes named "incertae sedis" or "environmental samples". The members of a container
aren't more closely related to one another than they are to the container's siblings;
the container is only present as a way to say something about the members. The fact
that a node had originally been in a container is recorded as a flag on the child
node.
When a subgenus X has the same name-string as its containing genus, its name-string
is changed to "X subgenus X". This follows a convention used by NCBI Taxonomy and
helps distinguish the two taxa later in assembly.
Sibling taxa with the same name-string are combined.
The normalized versions of the taxonomies then become the input to subsequent processing
phases.
Aligning nodes across taxonomies
This section and the next give details of the taxonomy combination method introduced
above.
OTT is assembled in a temporary work area or workspace by alternately aligning a source
to the workspace and merging that source into the workspace. It is important that
source taxonomy nodes be matched with workspace nodes when and only when this is appropriate.
A mistaken identity between a source node and a workspace node can be disastrous,
leading not just to an incorrect classification but to downstream curation errors
in OTU matching (e.g. putting a snail in flatworms). A mistaken non-identity (separation)
can also be a problem, since taxon duplication (i.e. multiple nodes for the same taxon)
leads to loss of unification opportunities in tree synthesis.
As described above, source taxonomies are processed (aligned and merged) in priority
order. For each source taxonomy, ad hoc adjustments are applied before automatic alignments.
For automatic alignment, alignments closest to the tips of the source taxonomy are
found in a first pass, and all others in a second pass. The two-pass structure permits
first-pass alignments to be used during the second pass (see Overlap, below).
Merging unaligned source nodes
After the alignment phase, we are left with the set of source nodes that could not
be aligned to the workspace. The next step is to determine if and how these (potentially
new) nodes can be merged into the workspace.
The combined taxonomy (U, above) is constructed by adding copies of unaligned nodes
from the source taxonomy S' one at a time to the workspace, which initially contains
a copy of S. Nodes of S' therefore correspond to workspace nodes in either of two
ways: by mapping to a copy of an S-node (via the S'-S alignment), or by mapping to
a copy of an S'-node (when there is no S'-S alignment for the S'-node).
As described above, each copied S'-node is part of either a graft or an insertion.
A graft or insertion rooted at r' is attached to the workspace as a child of the nearest
common ancestor node of r''s siblings' images. A graft is flagged incertae sedis if
that NCA is a node other than the parent of the sibling images. By construction, insertions
never have this property, so an insertion is never flagged incertae sedis.
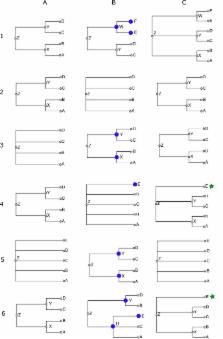
The following schematic examples illustrate each of the cases that come up while merging
taxonomies. Taxonomy fragments are written in Newick notation (Olsen 1990). Fig. 3
illustrates each of these six cases.
Case 1: ((a,b)x,(c,d)y)z + ((c,d)y,(e,f)w)z = ((a,b)x,(c,d)y,(e,f)w)z
This is a simple graft. The taxon w does not occur in the workspace, so it and its
children are copied. The workspace copy of w is attached as a sibling of its siblings'
images: its sibling is y in S', which is aligned to y in the workspace, so the copy
becomes a child of y's parent, or z.
Case 2: ((a,b)x,(c,d)y)z + (a,b,c,d)z = ((a,b)x,(c,d)y)z
No nodes are copied from S' to the workspace because every node in S' is aligned to
some node in S - there are no nodes that could be copied.
Case 3:(a,b,c,d)z + ((a,b)x,(c,d)y)z = ((a,b)x,(c,d)y)z
Supposing x and y are unaligned, then x and y from S' insert into the classification
of z. The workspace gets copies of these two S'-nodes.
Example: superfamily
Chitonoidea
, which is in WoRMS (S') but not in NCBI Taxonomy (S), inserts into NCBI Taxonomy.
Its parent is suborder
Chitonina
, which is in NCBI (i.e. aligned to the workspace), and its children are six families
that are all in NCBI (aligned).
Case 4: ((a,b)x,(c,d)y)z + (a,b,c,d,e)z = ((a,b)x,(c,d)y,?e)z
In this situation, we don't know where to put the unaligned taxon e from S': in x,
in y, or in z (sibling to x and y). The solution used here is to add e to z and mark
it as incertae sedis (indicated above by the question mark).
For example, family
Melyridae
from GBIF has five genera, of which two (
Trichoceble
,
Danacaea
) are not found in the workspace, and the other three do not all have the same parent
after alignment - they are in three different subfamilies.
Trichoceble
and
Danacaea
are made to be incertae sedis children of
Melyridae
, because there is no telling which NCBI subfamily they are supposed to go in.
Case 5: (a,b,c,d,e)z + ((a,b)x,(c,d)y)z = (a,b,c,d,e)z
We don't want to lose the fact from the higher priority taxonomy S that e is a proper
child of z (i.e. not incertae sedis), so we discard nodes x and y, ignoring what would
otherwise have been an insertion.
So that we have a term for this situation, say that x is absorbed into z.
Case 6: ((a,b)x,(c,d)y)z + ((a,c)p,(b,d,e)q)z = ((a,b)x,(c,d)y,?e)z
If the source has a hierarchy that is incompatible with the one in the workspace,
the conflicting source nodes are ignored, and any unaligned nodes (e) become incertae
sedis nodes under an ancestor containing the incompatible node's children.
For example, when WoRMS is merged, the workspace has, from NCBI,
((
Archaeognatha
)Monocondylia,(
Pterygota
,
Zygentoma
)
Dicondylia
)
Insecta
and the classification given by WoRMS is
((
Archaeognatha
,
Thysanura
=
Zygentoma
)Apteryogota,
Pterygota
)
Insecta
That is, NCBI groups
Thysanura
(
Zygentoma
) with
Pterygota
, while WoRMS groups it with
Archaeognatha
. The WoRMS hierarchy is ignored in favor of the higher priority NCBI hierarchy. If
Insecta
in WoRMS had had an unaligned third child, it would have ended up incertae sedis in
Insecta
.
The test for compatibility is very simple: a source node is incompatible with the
workspace if the nodes that its aligned children align with do not all have the same
parent.
Final patches
After all source taxonomies are aligned and merged, we apply general ad hoc additions
and patches to the workspace, in a manner similar to that employed with the source
taxonomies. Patches are represented in three formats. An early patch system used hand-written
tabular files, additions via the user interface use a machine-processed JSON format,
and most other patches are written as simple Python statements. There are 106 additions
in JSON form, 97 additions and patches in tabular form, and approximately 121 in Python
form.
Assigning identifiers
The final step is to assign unique, stable identifiers to nodes so that external links
to OTT nodes will continue to function correctly after the previous OTT version is
replaced by the new one.
Identifier assignment is done by aligning the previous version of OTT to the new version.
As with the other alignments, there are scripted ad hoc adjustments to correct for
some errors that would otherwise be made by automated assignment. For this alignment,
the set of heuristics is extended by adding rules that prefer candidates that have
the same source taxonomy node id as the previous version node being aligned. After
transferring identifiers of aligned nodes, any remaining workspace nodes are given
newly 'minted' identifiers.
The alignment is computed only for the purpose of assigning identifiers; the previous
OTT version is not merged into the workspace. An identifier can only persist from
one OTT version to the next if it continues to occur in some source taxonomy.
Results
The assembly method described above yields the reference taxonomy that is used by
the Open Tree of Life project. The taxonomy itself, the details of how the assembly
method unrolls to generate the taxonomy, and the degree to which the taxonomy meets
the goals set out for it are all of interest in assessing how, and how well, the method
works. We will address each of these three aspects of the method in turn.
Summary of Open Tree Taxonomy
The methods and results presented here are for version 3.0 of the Open Tree Taxonomy
(which follows five previous releases using the automated assembly method). The taxonomy
contains 3,594,550 total taxa; 3,272,177 tips; and 277,365 internal nodes. 2,335,412
of the nodes have a Linnean binomial of the form Genus epithet. There are 1,842,403
synonym records and 9,089 name-strings that are primary for more than one nodes. A
longer list of metrics is in Table 2.
The number of species level homonym name-strings (2867) is surprisingly high. While
a small number of these are legitimate, e.g.
Scoparia
dulci
which is used in practice for both a plant and an insect, most of them result from
errors in sources. Some originate in a single source, but most seem to be between
nodes contributed by multiple sources. We researched ten cases chosen at random, and
in every one, two sources disagreed on placement in separation taxa, and only one
source is correct. E.g.
Callirhynchius
exquisitus
is in beetles in one source and in decapods in another, so the Separation heuristic
prevents alignment, and the workspace ends up with two nodes. But the decapod placement
is incorrect, and there is really only one species. (Amazingly, every one of the ten
samples was later corrected in the source database!)
Results of assembly procedure
As OTT is assembled, the alignment procedure processes every source node, either choosing
an alignment target for it in the workspace based on the results of the heuristics,
or leaving it unaligned. Fig. 4 illustrates the action of the alignment phase. The
presence of a single candidate node does not automatically align the two nodes - we
still apply the heuristics to ensure a match (and occasionally reject the single candidate).
We counted the frequency of success for each heuristic, i.e. the number of times that
a particular heuristic was the one that accepted the winning candidate from among
two or more candidates. Table 3 shows these results. Separation (do not align taxa
in disjoint separation taxa; used first), Lineage (align taxa with shared lineage;
used midway through) and Same-name-string (prefer candidates who primary name-string
matches; used last) were by far the most frequent.
After assembly, the next step in the method is to merge the unaligned nodes into the
workspace taxonomy. Of the 3,780,949 unaligned nodes, the vast majority (99%) are
grafted into the workspace. The remaining nodes (<1%) are either insertions, absorptions
or remain unmerged due to ambiguities.
We also examined the fate of nodes from each of the input taxonomies, and Table 4
provides these results. The results are dependent on the order in which sources are
added to the workspace. Overall, the number of conflicts is relatively low (<1%).
Evaluating the taxonomy relative to requirements
The introduction sets out requirements for an Open Tree taxonomy. How well are these
requirements met?
Discussion
The primary actionable information in the source taxonomies consists of name-strings,
and therefore the core of our method is a set of heuristics that can handle the common
problems encountered when trying to merge hierarchies of name-strings. These problems
include expected taxonomic issues such as synonyms, homonyms, and differences in placement
and membership between sources. They also include errors such as duplications, spelling
mistakes, and misplaced taxa. The problem cases add up to over 100,000 difficult alignments
when the total number of source records measures over 6 million.
Ultimately there is no fully automated and foolproof test to determine whether two
nodes can be aligned - whether node A and node B, from different source taxonomies,
are about the same taxon. The information to do this is in the literature and in databases
on the Internet, but often it is (understandably) missing from the source taxonomies.
It is not feasible to investigate such problems individually, so the taxonomy assembly
methods identify and handle thousands of 'special cases' in an automated way. We currently
use only name-strings, rudimentary classification information, and (minimally) ranks
to guide assembly. We note the large role that our hand-curated "separation taxonomy"
played in the alignment phase. This is a set of taxa that are consistent across the
various sources, and allow us to make the (seemingly obvious) determination "these
two taxa are in completely separate groups, so do not align them".
Community curation
We have also developed a system for curators to directly add new taxon records to
the taxonomy from published phylogenies, which often contain newly described species
that are not yet present in any source taxonomy. These taxon records include provenance
information, including references explaining the taxon, and the identity of the curator.
We expose this provenance information through the web site and the taxonomy API.
We also provide a feedback mechanism on the synthetic tree browser, and find that
most of the comments left are about presence, absence, choice, and spelling of labels,
rather than the topology of the synthetic tree. These are issues that are addressed
by improvements to the taxonomy. Expanding this feature to capture this feedback in
a more structured, and therefore machine-readable, format would allow users to directly
contribute taxonomic patches to the system.
Comparison to other taxonomies
Given the unique goals of the Open Tree Taxonomy in comparison to most other taxonomy
projects, it is difficult to compare OTT to other taxonomies in a meaningful way.
The Open Tree Taxonomy is technically most similar to the GBIF taxonomy, in the sense
that each is a synthesis of existing, overlapping taxonomies rather than a curated
taxonomic database or one based on grafting. The GBIF method is yet unpublished (for
basic information on the GBIF backbone see Döring 2016a, Döring 2016b). Once the GBIF
method has been formally described, it will be useful to compare the two approaches
and identify common and unique techniques for automated, scalable name-string matching
and hierarchy merging.
Potential improvements and future work
The development of the assembly software has been driven by the needs of the Open
Tree project, not by any concerted effort to create a widely applicable or theoretically
principled tool. A system like this is never finished, and this one is in its infancy.
There are endless opportunities for bringing additional techniques, methods, data,
and code libraries to bear, and we have faced difficult choices in deciding where
to put our effort. Following are some of the directions for development that could
have the highest impact.
It is likely that alignment and merge could be improved by making better use of species
proximity implied by the shape of the classification, and decreasing its reliance
on the names of internal nodes. Better use of proximity might permit separation and
identification of tips without use of a separation taxonomy, removing the need for
the manual work of maintaining the separation taxonomy and the adjustment directives
needed to align source taxonomies to it. An example is
Conolophus
, where two genus-level nodes in the same separation taxon are mistakenly combined.
How to accomplish this is not obvious, but it is not obviously impossible.
The alignment method should be extended to make use of authority information, when
it is available. If name-strings match, or even if just species epithets match, then
matching authority information is good evidence that the same taxon is meant. The
form of authority information varies between sources, but could be normalized using
the Global Names Parser (Patterson et al. 2016).
Name-strings could also be analyzed to detect partial matches, e.g. matching on species
epithets even when the genus disagrees, and spelling and gender variant recognition.
Doing so would eliminate thousands of duplications. Other work on name matching, such
as the Global Names Resolver (Patterson et al. 2016), goes far beyond what is done
for OTT and these techniques should be used.
The redescription problem described above should be addressed to the extent possible.
Our handling of duplicate records in source taxonomies is incomplete and needs to
be fixed. If two source nodes can be aligned to the same workspace node, then the
duplication will not affect the workspace. But if there is no workspace node to align
them to, the duplication persists in the workspace after the source taxonomy has been
merged. There is special case logic when taxonomies are imported to fold together
sibling duplicates, but not alignable duplicates generally.
An assembly run can lead to a variety of error conditions and test failures. Currently
these are difficult to diagnose, mainly for lack of technology for displaying the
particular pieces of the sources, workspace, and assembly history that are relevant
to the error. Once this information is surfaced it is usually not too difficult to
work out a fix in the form of a patch or an improvement to the program logic. A small
amount of automation could speed this kind of investigation and save curator time.
The community curation should be developed, as mentioned above. Its success would
depend on allowing users to test proposed changes and diagnose and repair any problems
with them.
Curators frequently request new taxonomy sources. The most frequently requested are
improved fish, bird, plant, and paleontological sources. Community members have also
suggested the Plazi TreatmentBank (Miller et al. 2015). Again, the information is
generally available, but not yet harvested. (Some frequently requested sources may
only be accessed under agreement with contractual terms (variously called "terms of
use" or a "data use agreement"). One of these is the IUCN Red List (International
Union for Conservation of Nature and Natural Resources 2016), an important source
of up-to-date information on mammal species. These sources are off limits to Open
Tree due to the project's open data requirement.)
The presence of invalid and unaccepted names remains a significant problem. The information
needed to detect them is available, and could be harvested.
Basic usability features for application to new projects would include proper packaging
of the application, and support for Darwin Core (Wieczorek et al. 2012) for both input
and output.
Future work on taxonomy aggregation should attempt a more rigorous and pluralistic
approach to classification (Franz et al. 2016a, Kennedy et al. 2006, Lepage et al.
2014, Franz et al. 2016b). Alignment should detect and record lumping and splitting
events, and the classification conflicts detected during merge should be exposed to
users. Exposing conflicts is in the interest of scientific transparency. Retaining
alternative groupings could be useful in phylogenetic analysis, as a check on which
of the sources agree or disagree with a given analysis. Lumping and splitting due
to redescription, which lead to the same name-string (including author) referring
to different taxa in different sources, could be recorded using multiple nodes qualified
by description or source ('sensu'). Ideally, better handling of descriptions in aggregators
ought to encourage sources to make links to primary sources more readily available
for a variety of purposes.
Data resources
All source code is open source (licensed BSD 2-clause) and available on GitHub at
https://github.com/OpenTreeOfLife/reference-taxonomy. A snapshot of the code used
to produce the version of OTT described here is archived at Zenodo (https://doi.org/10.5281/zenodo.546111).
All data, including Open Tree Taxonomy 3.0 and all processed source taxonomies is
archived on Dryad (cannot be uploaded before paper accepted; version for review at
https://github.com/OpenTreeOfLife/reference-taxonomy/tree/master/doc/method/data-package).
Conclusions
We have presented a method for merging multiple taxonomies into a single synthetic
taxonomy. The method is designed to produce a taxonomy optimized for the Open Tree
of Life phylogenetic tree synthesis project. Most taxonomy projects are databases
of taxonomy information that are continuously updated by curators as new information
is published in the taxonomic literature. In contrast, the Open Tree Taxonomy takes
several of these curated taxonomies and assembles a synthetic taxonomy de novo each
time a new version of the taxonomy is needed.
We have also developed a system for curators to directly add new taxa to the taxonomy
from published phylogenies. These taxon additions include provenance information,
including the source of the taxon and identity of the curator. We expose this provenance
information through the website and the taxonomy API. Most of the Open Tree feedback
has been about taxonomy, and expanding this feature to other types of taxonomic information
allows users to directly contribute expertise and allows projects to easily share
that information.
Taxonomic information is certainly best curated at a scale smaller than "all life"
by experts in a particular group. Therefore, producing comprehensive taxonomies is
always a synthesis of curated taxonomies. We advocate for the type of methods being
used by Open Tree and by GBIF, where synthesis of overlapping sources is done in a
repeatable fashion from sources, allowing changed information in sources to be quickly
included in the comprehensive taxonomy, and also allowing continuous improvement to
the synthesis method. Provenance information is retained and presented as part of
the synthetic taxonomy. This type of synthesis requires that source taxonomies be
available online, either through APIs or by bulk download, in a format that can be
easily parsed, and ideally without terms of use that prevent distribution and reuse
of the resulting synthetic taxonomies.
Related collections
Most cited references27

- Record: found
- Abstract: found
- Article: found
Towards resolving Lamiales relationships: insights from rapidly evolving chloroplast sequences
- Record: found
- Abstract: not found
- Article: not found
Research and Societal Benefits of the Global Biodiversity Information Facility
JAMES L. EDWARDS (2004)
- Record: found
- Abstract: found
- Article: found
Avibase – a database system for managing and organizing taxonomic concepts
Denis Lepage, Gaurav Vaidya, Robert Guralnick (2014)
Author and article information
Comments
Comment on this article
scite_
0
0
0
0
 Smart Citations
Smart Citations0
0
0
0
Citing PublicationsSupportingMentioningContrasting
See how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.