- Record: found
- Abstract: found
- Article: found
Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids
Read this article at
Abstract
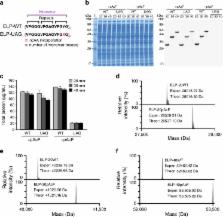
Cell-free protein synthesis has emerged as a powerful approach for expanding the range of genetically encoded chemistry into proteins. Unfortunately, efforts to site-specifically incorporate multiple non-canonical amino acids into proteins using crude extract-based cell-free systems have been limited by release factor 1 competition. Here we address this limitation by establishing a bacterial cell-free protein synthesis platform based on genomically recoded Escherichia coli lacking release factor 1. This platform was developed by exploiting multiplex genome engineering to enhance extract performance by functionally inactivating negative effectors. Our most productive cell extracts enabled synthesis of 1,780 ± 30 mg/L superfolder green fluorescent protein. Using an optimized platform, we demonstrated the ability to introduce 40 identical p-acetyl- l-phenylalanine residues site specifically into an elastin-like polypeptide with high accuracy of incorporation ( ≥ 98%) and yield (96 ± 3 mg/L). We expect this cell-free platform to facilitate fundamental understanding and enable manufacturing paradigms for proteins with new and diverse chemistries.
Abstract
Cell-free protein synthesis allows for producing proteins without the need of a host organism, thus sparing the researcher experimental hassle. Here, the authors developed a cell-free synthesis method that enables incorporating non-standard amino acids in the product.
Related collections
Most cited references54
- Record: found
- Abstract: found
- Article: not found
Adding new chemistries to the genetic code.
- Record: found
- Abstract: not found
- Article: not found
Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins.
- Record: found
- Abstract: found
- Article: not found