- Record: found
- Abstract: found
- Article: found
Evolution of the Transmission-Blocking Vaccine Candidates Pvs28 and Pvs25 in Plasmodium vivax: Geographic Differentiation and Evidence of Positive Selection
Read this article at
Abstract
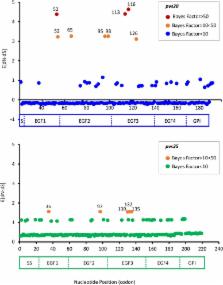
Transmission-blocking (TB) vaccines are considered an important tool for malaria control and elimination. Among all the antigens characterized as TB vaccines against Plasmodium vivax, the ookinete surface proteins Pvs28 and Pvs25 are leading candidates. These proteins likely originated by a gene duplication event that took place before the radiation of the known Plasmodium species to primates. We report an evolutionary genetic analysis of a worldwide sample of pvs28 and pvs25 alleles. Our results show that both genes display low levels of genetic polymorphism when compared to the merozoite surface antigens AMA-1 and MSP-1; however, both ookinete antigens can be as polymorphic as other merozoite antigens such as MSP-8 and MSP-10. We found that parasite populations in Asia and the Americas are geographically differentiated with comparable levels of genetic diversity and specific amino acid replacements found only in the Americas. Furthermore, the observed variation was mainly accumulated in the EGF2- and EGF3-like domains for P. vivax in both proteins. This pattern was shared by other closely related non-human primate parasites such as Plasmodium cynomolgi, suggesting that it could be functionally important. In addition, examination with a suite of evolutionary genetic analyses indicated that the observed patterns are consistent with positive natural selection acting on Pvs28 and Pvs25 polymorphisms. The geographic pattern of genetic differentiation and the evidence for positive selection strongly suggest that the functional consequences of the observed polymorphism should be evaluated during development of TBVs that include Pvs25 and Pvs28.
Author Summary
Plasmodium vivax is the most prevalent human malarial parasite outside Africa. The fact that patients can relapse due to the parasite dormant liver stages, among other biologic and epidemiologic characteristics of vivax malaria, facilitates the persistence of the disease in many endemic areas. These challenges have fueled the search for new control tools, including transmission blocking (TB) vaccines targeting the parasite sexual stages. Here we study the genetic diversity of two major TB vaccine antigens, Pvs25 and Pvs28. We show that these genes are relatively conserved worldwide but still harbor diversity that is not evenly distributed across the genes. These patterns are shared by the same proteins in closely related parasite species suggesting their functional importance. We also identify strong geographic differentiation between the circulating variants found in Asia and the Americas. Finally, evolutionary genetic analyses indicate that the observed variation in both genes could be maintained by natural selection. Thus, these polymorphisms may confer an adaptive advantage to the parasite. These results indicate that the genetic variation found in these genes and their geographic distribution should be considered by vaccine developers.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
DnaSP, DNA polymorphism analyses by the coalescent and other methods.
- Record: found
- Abstract: found
- Article: not found
Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology.
- Record: found
- Abstract: found
- Article: not found