- Record: found
- Abstract: found
- Article: found
Subunits of an E3 Ligase Complex as Degrons for Efficient Degradation of Cytosolic, Nuclear, and Membrane Proteins

Read this article at
Abstract
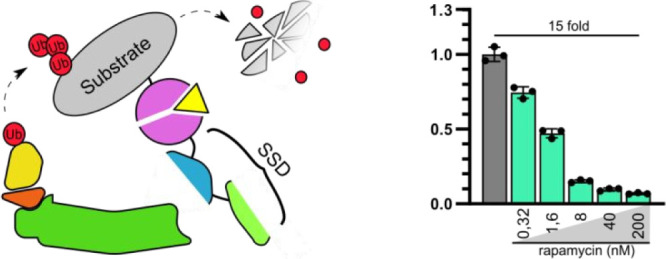
Protein degradation is a highly regulated cellular process crucial to enable the high dynamic range of the response to external and internal stimuli and to balance protein biosynthesis to maintain cell homeostasis. Within mammalian cells, hundreds of E3 ubiquitin ligases target specific protein substrates and could be repurposed for synthetic biology. Here, we present a systematic analysis of the four protein subunits of the multiprotein E3 ligase complex as scaffolds for the designed degrons. While all of them were functional, the fusion of a fragment of Skp1 with the target protein enabled the most effective degradation. Combination with heterodimerizing peptides, protease substrate sites, and chemically inducible dimerizers enabled the regulation of protein degradation. While the investigated subunits of E3 ligases showed variable degradation efficiency of the membrane and cytosolic and nuclear proteins, the bipartite SSD (SOCSbox-Skp1(ΔC111)) degron enabled fast degradation of protein targets in all tested cellular compartments, including the nucleus and plasma membrane, in different cell lines and could be chemically regulated. These subunits could be employed for research as well as for diverse applications, as demonstrated in the regulation of Cas9 and chimeric antigen receptor proteins.
Related collections
Most cited references76
- Record: found
- Abstract: not found
- Article: not found
Enzymatic assembly of DNA molecules up to several hundred kilobases.
- Record: found
- Abstract: found
- Article: not found
CRISPR RNA-guided activation of endogenous human genes
- Record: found
- Abstract: found
- Article: not found