- Record: found
- Abstract: found
- Article: found
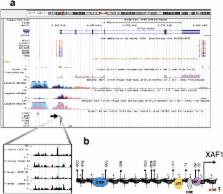
Epigenetic silencing of the XAF1 gene is mediated by the loss of CTCF binding
Read this article at
Abstract
XAF1 is a tumour suppressor gene that compromises cell viability by modulating different cellular events such as mitosis, cell cycle progression and apoptosis. In cancer, the XAF1 gene is commonly silenced by CpG-dinucleotide hypermethylation of its promoter. DNA demethylating agents induce transcriptional reactivation of XAF1, sensitizing cancer cells to therapy. The molecular mechanisms that mediate promoter CpG methylation have not been previously studied. Here, we demonstrate that CTCF interacts with the XAF1 promoter in vivo in a methylation-sensitive manner. By transgene assays, we demonstrate that CTCF mediates the open-chromatin configuration of the XAF1 promoter, inhibiting both CpG-dinucleotide methylation and repressive histone posttranslational modifications. In addition, the absence of CTCF in the XAF1 promoter inhibits transcriptional activation induced by well-known apoptosis activators. We report for the first time that epigenetic silencing of the XAF1 gene is a consequence of the loss of CTCF binding.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
DNA hypermethylation in tumorigenesis: epigenetics joins genetics.
- Record: found
- Abstract: found
- Article: not found
CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species.
- Record: found
- Abstract: found
- Article: not found