- Record: found
- Abstract: found
- Article: found
Characteristics of Microbial Communities in Crustal Fluids in a Deep-Sea Hydrothermal Field of the Suiyo Seamount
Read this article at
Abstract
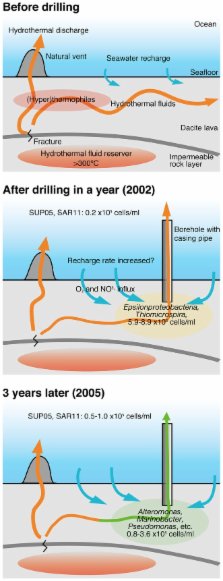
To directly access the sub-seafloor microbial communities, seafloor drilling has been done in a deep-sea hydrothermal field of the Suiyo Seamount, Izu-Bonin Arc, Western Pacific. In the present study, crustal fluids were collected from the boreholes, and the bacterial and archaeal communities in the fluids were investigated by culture-independent molecular analysis based on 16S rRNA gene sequences. Bottom seawater, sands, rocks, sulfide mound, and chimneys were also collected around the boreholes and analyzed for comparisons. Comprehensive analysis revealed the characteristics of the microbial community composition in the crustal fluids. Phylotypes closely related to cultured species, e.g., Alteromonas, Halomonas, Marinobacter, were relatively abundant in some crustal fluid samples, whereas the phylotypes related to Pelagibacter and the SUP05-group were relatively abundant in the seawater samples. Phylotypes related to other uncultured environmental clones in Alphaproteobacteria and Gammaproteobacteria were relatively abundant in the sand, rock, sulfide mound, and chimney samples. Furthermore, comparative analysis with previous studies of the Suiyo Seamount crustal fluids indicates the change in the microbial community composition for 3 years. Our results provide novel insights into the characteristics of the microbial communities in crustal fluids beneath a deep-sea hydrothermal field.
Related collections
Most cited references30
- Record: found
- Abstract: found
- Article: not found
Archaea in coastal marine environments.
- Record: found
- Abstract: found
- Article: not found
Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data
- Record: found
- Abstract: found
- Article: not found