- Record: found
- Abstract: found
- Article: not found
Ecology, evolution and spillover of coronaviruses from bats
Abstract
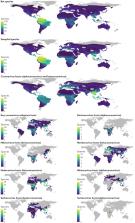
In the past two decades, three coronaviruses with ancestral origins in bats have emerged and caused widespread outbreaks in humans, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since the first SARS epidemic in 2002–2003, the appreciation of bats as key hosts of zoonotic coronaviruses has advanced rapidly. More than 4,000 coronavirus sequences from 14 bat families have been identified, yet the true diversity of bat coronaviruses is probably much greater. Given that bats are the likely evolutionary source for several human coronaviruses, including strains that cause mild upper respiratory tract disease, their role in historic and future pandemics requires ongoing investigation. We review and integrate information on bat–coronavirus interactions at the molecular, tissue, host and population levels. We identify critical gaps in knowledge of bat coronaviruses, which relate to spillover and pandemic risk, including the pathways to zoonotic spillover, the infection dynamics within bat reservoir hosts, the role of prior adaptation in intermediate hosts for zoonotic transmission and the viral genotypes or traits that predict zoonotic capacity and pandemic potential. Filling these knowledge gaps may help prevent the next pandemic.
Abstract
Bats harbour a multitude of coronaviruses and owing to their diversity and wide distribution are prime reservoir hosts of emerging viruses. Ruiz-Aravena, McKee and colleagues analyse the currently available information on bat coronaviruses and discuss their role in recent and potential future spillovers.
Related collections
Most cited references173
- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin

- Record: found
- Abstract: found
- Article: found
Re-epithelialization and immune cell behaviour in an ex vivo human skin model
- Record: found
- Abstract: found
- Article: not found
