- Record: found
- Abstract: found
- Article: found
scBridge embraces cell heterogeneity in single-cell RNA-seq and ATAC-seq data integration

Read this article at
Abstract
Single-cell multi-omics data integration aims to reduce the omics difference while keeping the cell type difference. However, it is daunting to model and distinguish the two differences due to cell heterogeneity. Namely, even cells of the same omics and type would have various features, making the two differences less significant. In this work, we reveal that instead of being an interference, cell heterogeneity could be exploited to improve data integration. Specifically, we observe that the omics difference varies in cells, and cells with smaller omics differences are easier to be integrated. Hence, unlike most existing works that homogeneously treat and integrate all cells, we propose a multi-omics data integration method (dubbed scBridge) that integrates cells in a heterogeneous manner. In brief, scBridge iterates between i) identifying reliable scATAC-seq cells that have smaller omics differences, and ii) integrating reliable scATAC-seq cells with scRNA-seq data to narrow the omics gap, thus benefiting the integration for the rest cells. Extensive experiments on seven multi-omics datasets demonstrate the superiority of scBridge compared with six representative baselines.
Abstract
Multi-omics data integration can be challenging in the event of cell heterogeneity. Here, the authors present scBridge, a method that exploits heterogeneous omics differences, to progressively integrate cells and narrows omics gap, leading to promising integration and label transfer results.
Related collections
Most cited references46
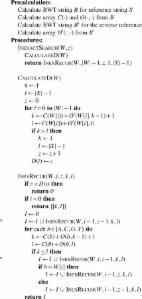
- Record: found
- Abstract: found
- Article: found
Fast and accurate short read alignment with Burrows–Wheeler transform
- Record: found
- Abstract: found
- Article: found
SciPy 1.0: fundamental algorithms for scientific computing in Python
- Record: found
- Abstract: found
- Article: not found