- Record: found
- Abstract: found
- Article: found
Ebola Virus Epidemiology, Transmission, and Evolution during Seven Months in Sierra Leone
Read this article at
Summary
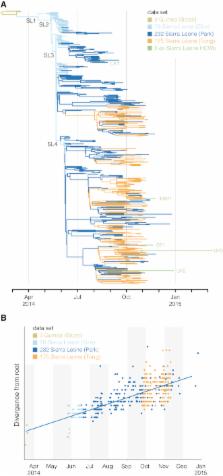
The 2013–2015 Ebola virus disease (EVD) epidemic is caused by the Makona variant of Ebola virus (EBOV). Early in the epidemic, genome sequencing provided insights into virus evolution and transmission and offered important information for outbreak response. Here, we analyze sequences from 232 patients sampled over 7 months in Sierra Leone, along with 86 previously released genomes from earlier in the epidemic. We confirm sustained human-to-human transmission within Sierra Leone and find no evidence for import or export of EBOV across national borders after its initial introduction. Using high-depth replicate sequencing, we observe both host-to-host transmission and recurrent emergence of intrahost genetic variants. We trace the increasing impact of purifying selection in suppressing the accumulation of nonsynonymous mutations over time. Finally, we note changes in the mucin-like domain of EBOV glycoprotein that merit further investigation. These findings clarify the movement of EBOV within the region and describe viral evolution during prolonged human-to-human transmission.
Graphical Abstract
Highlights
-
•
In Sierra Leone, transmission has primarily been within-country, not between-country
-
•
Infectious doses are large enough for intrahost variants to transmit between hosts
-
•
A prolonged epidemic removes deleterious mutations from the viral population
-
•
There is preliminary evidence for human RNA editing effects on the Ebola genome
Abstract
Ebola virus genomes from 232 patients sampled over 7 months in Sierra Leone were sequenced. Transmission of intrahost genetic variants suggests a sufficiently high infectious dose during transmission. The human host may have caused direct alterations to the Ebola virus genome.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Time dependency of molecular rate estimates and systematic overestimation of recent divergence times.
- Record: found
- Abstract: found
- Article: not found