- Record: found
- Abstract: found
- Article: found
Characterizing the “fungal shunt”: Parasitic fungi on diatoms affect carbon flow and bacterial communities in aquatic microbial food webs
Read this article at
Significance
Planktonic microorganisms interact with each other in multifarious ways, ultimately catalyzing the flow of carbon and energy in diverse aquatic environments. However, crucial links associated with eukaryotic microparasites are still overlooked in planktonic networks. We addressed such links by studying cryptic interactions between parasitic fungi, phytoplankton, and bacteria using a model pathosystem. Our results demonstrate that parasitic fungi profoundly modified microbial interactions through several mechanisms (e.g., transferring photosynthetic carbon to infecting fungi, stimulating bacterial colonization on phytoplankton cells, and altering the community composition of bacteria and their acquisition of photosynthetic carbon). Hence, fungal microparasites can substantially shape the microbially mediated carbon flow at the base of aquatic food webs and should be considered as crucial members within plankton communities.
Abstract
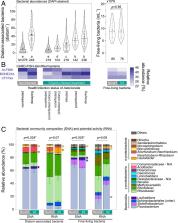
Microbial interactions in aquatic environments profoundly affect global biogeochemical cycles, but the role of microparasites has been largely overlooked. Using a model pathosystem, we studied hitherto cryptic interactions between microparasitic fungi (chytrid Rhizophydiales), their diatom host Asterionella, and cell-associated and free-living bacteria. We analyzed the effect of fungal infections on microbial abundances, bacterial taxonomy, cell-to-cell carbon transfer, and cell-specific nitrate-based growth using microscopy (e.g., fluorescence in situ hybridization), 16S rRNA gene amplicon sequencing, and secondary ion mass spectrometry. Bacterial abundances were 2 to 4 times higher on individual fungal-infected diatoms compared to healthy diatoms, particularly involving Burkholderiales. Furthermore, taxonomic compositions of both diatom-associated and free-living bacteria were significantly different between noninfected and fungal-infected cocultures. The fungal microparasite, including diatom-associated sporangia and free-swimming zoospores, derived ∼100% of their carbon content from the diatom. By comparison, transfer efficiencies of photosynthetic carbon were lower to diatom-associated bacteria (67 to 98%), with a high cell-to-cell variability, and even lower to free-living bacteria (32%). Likewise, nitrate-based growth for the diatom and fungi was synchronized and faster than for diatom-associated and free-living bacteria. In a natural lacustrine system, where infection prevalence reached 54%, we calculated that 20% of the total diatom-derived photosynthetic carbon was shunted to the parasitic fungi, which can be grazed by zooplankton, thereby accelerating carbon transfer to higher trophic levels and bypassing the microbial loop. The herein termed “fungal shunt” can thus significantly modify the fate of photosynthetic carbon and the nature of phytoplankton–bacteria interactions, with implications for diverse pelagic food webs and global biogeochemical cycles.
Related collections
Most cited references105
- Record: found
- Abstract: found
- Article: not found
Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples.
- Record: found
- Abstract: found
- Article: not found
A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life
- Record: found
- Abstract: found
- Article: not found