- Record: found
- Abstract: found
- Article: found
Potentiation of B2 receptor signaling by AltB2R, a newly identified alternative protein encoded in the human bradykinin B2 receptor gene
Read this article at
Abstract
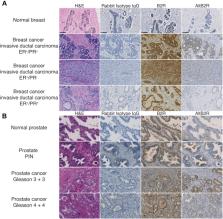
Recent functional and proteomic studies in eukaryotes ( www.openprot.org) predict the translation of alternative open reading frames (AltORFs) in mature G-protein-coupled receptor (GPCR) mRNAs, including that of bradykinin B2 receptor (B2R). Our main objective was to determine the implication of a newly discovered AltORF resulting protein, termed AltB2R, in the known signaling properties of B2R using complementary methodological approaches. When ectopically expressed in HeLa cells, AltB2R presented predominant punctate cytoplasmic/perinuclear distribution and apparent cointeraction with B2R at plasma and endosomal/vesicular membranes. The presence of AltB2R increases intracellular [Ca 2+] and ERK1/2-MAPK activation ( via phosphorylation) following B2R stimulation. Moreover, HEK293A cells expressing mutant B2R lacking concomitant expression of AltB2R displayed significantly decreased maximal responses in agonist-stimulated Gα q-Gα i2/3–protein coupling, IP 3 generation, and ERK1/2-MAPK activation as compared with wild-type controls. Conversely, there was no difference in cell-surface density as well as ligand-binding properties of B2R and in efficiencies of cognate agonists at promoting B2R internalization and β-arrestin 2 recruitment. Importantly, both AltB2R and B2R proteins were overexpressed in prostate and breast cancers, compared with their normal counterparts suggesting new associative roles of AltB2R in these diseases. Our study shows that BDKRB2 is a dual-coding gene and identifies AltB2R as a novel positive modulator of some B2R signaling pathways. More broadly, it also supports a new, unexpected alternative proteome for GPCRs, which opens new frontiers in fields of GPCR biology, diseases, and drug discovery.
Related collections
Most cited references59
- Record: found
- Abstract: found
- Article: not found
Trends in GPCR drug discovery: new agents, targets and indications
- Record: found
- Abstract: found
- Article: not found
Proteogenomics: concepts, applications and computational strategies.
- Record: found
- Abstract: found
- Article: not found