- Record: found
- Abstract: found
- Article: found
Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community
Read this article at
Abstract
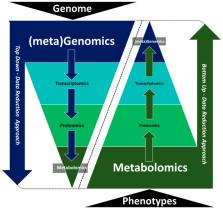
The use of multiple omics techniques (i.e., genomics, transcriptomics, proteomics, and metabolomics) is becoming increasingly popular in all facets of life science. Omics techniques provide a more holistic molecular perspective of studied biological systems compared to traditional approaches. However, due to their inherent data differences, integrating multiple omics platforms remains an ongoing challenge for many researchers. As metabolites represent the downstream products of multiple interactions between genes, transcripts, and proteins, metabolomics, the tools and approaches routinely used in this field could assist with the integration of these complex multi-omics data sets. The question is, how? Here we provide some answers (in terms of methods, software tools and databases) along with a variety of recommendations and a list of continuing challenges as identified during a peer session on multi-omics integration that was held at the recent ‘Australian and New Zealand Metabolomics Conference’ (ANZMET 2018) in Auckland, New Zealand (Sept. 2018). We envisage that this document will serve as a guide to metabolomics researchers and other members of the community wishing to perform multi-omics studies. We also believe that these ideas may allow the full promise of integrated multi-omics research and, ultimately, of systems biology to be realized.
Related collections
Most cited references178
- Record: found
- Abstract: found
- Article: found
The FAIR Guiding Principles for scientific data management and stewardship
- Record: found
- Abstract: found
- Article: not found
Causal analysis approaches in Ingenuity Pathway Analysis
- Record: found
- Abstract: found
- Article: not found