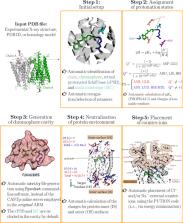
Laura Pedraza-González:
ORCID: http://orcid.org/0000-0002-9806-236X
laura.pedraza@unisi.it
Leonardo Barneschi:
ORCID: http://orcid.org/0000-0003-0736-1696
leonardo.barneschi@student.unisi.it
Daniele Padula:
ORCID: http://orcid.org/0000-0002-7171-7928
daniele.padula@unisi.it
Luca De Vico:
ORCID: http://orcid.org/0000-0002-2821-5711
Luca.DeVico@unisi.it
Massimo Olivucci:
ORCID: http://orcid.org/0000-0002-8247-209X
olivucci@unisi.it
Journal ID (nlm-ta): Top Curr Chem (Cham)
Journal ID (iso-abbrev): Top Curr Chem (Cham)
Title:
Topics in Current Chemistry (Cham)
Publisher:
Springer International Publishing
(Cham
)
ISSN
(Print):
2365-0869
ISSN
(Electronic):
2364-8961
Publication date
(Electronic):
15
March
2022
Publication date PMC-release: 15
March
2022
Publication date
(Print):
2022
Volume: 380
Issue: 3
Electronic Location Identifier: 21
Affiliations
[1
]GRID grid.9024.f, ISNI 0000 0004 1757 4641, Dipartimento di Biotecnologie, Chimica e Farmacia, , Università degli Studi di Siena, ; Via Aldo Moro 2, 53100 Siena, Italy
[2
]GRID grid.253248.a, ISNI 0000 0001 0661 0035, Department of Chemistry, , Bowling Green State University, ; Bowling Green, OH 43403 USA
[3
]GRID grid.5395.a, ISNI 0000 0004 1757 3729, Present Address: Department of Chemistry and Industrial Chemistry, , University of Pisa, ; Via Moruzzi 13, 56124 Pisa, Italy
Author information
Article
Publisher ID:
374
DOI: 10.1007/s41061-022-00374-w
PMC ID: 8924150
PubMed ID: 35291019
SO-VID: 2e4bec7d-ec32-49fa-af6d-cd72278c0197
Copyright © © The Author(s) 2022
License:
Open AccessThis article is licensed under a Creative Commons Attribution 4.0 International License,
which permits use, sharing, adaptation, distribution and reproduction in any medium
or format, as long as you give appropriate credit to the original author(s) and the
source, provide a link to the Creative Commons licence, and indicate if changes were
made. The images or other third party material in this article are included in the
article's Creative Commons licence, unless indicated otherwise in a credit line to
the material. If material is not included in the article's Creative Commons licence
and your intended use is not permitted by statutory regulation or exceeds the permitted
use, you will need to obtain permission directly from the copyright holder. To view
a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/.
Funded by: FundRef http://dx.doi.org/10.13039/100000001, National Science Foundation;
Award ID: CHE-CLP-1710191
Award Recipient
:
Massimo Olivucci
Funded by: FundRef http://dx.doi.org/10.13039/100000002, National Institutes of Health;
Award ID: 1R15GM126627 01
Award Recipient
:
Massimo Olivucci
Funded by: FundRef http://dx.doi.org/10.13039/501100003407, Ministero dell’Istruzione, dell’Università e della Ricerca;
Award ID: Rita Levi Montalcini grant, 2021-2024
Award Recipient
:
Daniele Padula